
TMEM43 promotes the development of hepatocellular carcinoma by activating VDAC1 through USP7 deubiquitination
Highlight box
Key findings
• In this study, we found for the first time that transmembrane protein 43 (TMEM43) is highly expressed in hepatocellular carcinoma (HCC). TMEM43 plays an important role in the occurrence and development of HCC. Further research has found an important regulatory relationship between TMEM43 and voltage-dependent anion channel 1 (VDAC1). In addition, we also found that ubiquitin-specific protease 7 (USP7) regulates the ubiquitination level of TMEM43 through ubiquitination, thereby affecting cancer progression.
What is known and what is new?
• TMEM43 is highly expressed in HCC and can regulate the proliferation, differentiation, apoptosis, and migration functions of HCC.
• Our research suggests that the USP7/TMEM43/VDAC1 signaling axis plays an important role in inhibiting cancer progression.
What is the implication, and what should change now?
• USP7/TMEM43/VDAC1 may serve as a regulatory molecular axis for HCC treatment. These findings may provide a new therapeutic target for HCC.
Introduction
On a global scale, hepatocellular carcinoma (HCC) is a common and deadly malignant tumor. In the United States, it ranks fifth in terms of mortality rate. Patients are usually diagnosed at an advanced stage, leading to poor prognosis (1-3). At present, surgical resection and liver transplantation are the most effective treatment methods, and the prognosis of cancer is poor. Only 5–15% of early-stage patients are eligible for surgical resection (4,5). Although there have been many studies on epigenetic changes, the molecular mechanism of HCC has not yet been fully understood (6,7). Therefore, it is necessary to further explore the potential molecular mechanisms of HCC and discover new potential targets to inhibit the development of HCC.
Transmembrane protein 43 (TMEM43), a member of the TMEM subfamily, is encoded by a highly conserved gene and is widely expressed in most species from bacteria to humans (8). Systematic genetic analysis has shown that TMEM43 is involved in metabolic related pathways (9). A previous study showed that TMEM43 regulates ferroptosis by reducing the expression of P53 (10). TMEM43 also plays an important role in tumors. A study has shown that the expression of TMEM43 is closely related to the clinical pathological characteristics and adverse survival outcomes of pancreatic cancer. In addition, they also found that inhibition of TMEM43 can inhibit the growth, migration, and invasion of pancreatic cancer in vitro and in vivo (11). However, the role of TMEM43 in cancer is not yet clear.
In this study, we first mined TMEM43 through the RNA sequencing (RNA-seq) and The Cancer Genome Atlas (TGCA) databases. Further research found that TMEM43 is highly expressed in HCC, and TMEM43 regulates the proliferation, cell cycle, apoptosis, and invasion of HCC through voltage-dependent anion channel 1 (VDAC1). Ubiquitin modification is an important post-translational modification process that regulates intracellular protein levels by adding ubiquitin molecules. This modification involves various cellular processes, including cell proliferation, apoptosis, invasion, migration, and DNA damage repair, which play crucial roles in tumor development and cancer progression (12). In addition, we found that ubiquitin-specific protease 7 (USP7) affected the progression of HCC by regulating the ubiquitination level of TMEM43 through deubiquitination. We present this article in accordance with the MDAR reporting checklist (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-108/rc).
Methods
Clinical samples
Tumor tissue samples were taken from 96 patients with primary HCC who underwent surgery in the Affiliated Hospital of Nantong University. This study was approved by the Institutional Ethics Committee of the Affiliated Hospital of Nantong University (ID: 2022-L062). Informed consent was provided by each participant. The study was conducted in accordance with the Declaration of Helsinki (revised in 2013).
Cell culture and transfection
HCC cell lines (Huh7, HCCLM3) were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). HCCLM3 cells are a type of hepatoma cells line with high metastatic potential (13,14). Hepatoma cells were cultured in Dulbecco’s modified Eagle medium (DMEM) medium containing 10% fetal bovine serum (FBS). Plasmids were transfected into cells using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) and transfection efficiency was evaluated using western blot (WB). The sequences of the short hairpin (sh) RNAs are as follows: sh-TMEM43#1, “CCTCAACCTTATGACACGGAT”; sh-TMEM43#2, “CCTTTGTCCAAATTGGCAGGT”; sh-TMEM43#3, “GAGATGTACCAATGGGTAGAA”; sh-USP7, “CGTGGTGTCAAGGTGTACTAA”.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using Trizol reagent (Invitrogen) and complementary DNA (cDNA) was obtained using Prime Script RT kit (Takara Bio, Shiga, Japan). Using DNA as a template, SYBR Select Master Mix kit (Invitrogen) and ABI Prism 7900 detection system (Applied Biosystems, Carlsbad, CA, USA) were used for qRT-PCR reaction. The primers for MiR-7 were designed and synthesized by RiboBio (Shanghai, China). The upstream primer article number of MiR-7 was SSD809232326 and the downstream primer article number was SSD089261711.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was detected using the CCK-8 method. After inoculating the cell suspension in a 96 well plate, 10 µL of CCK-8 solution was added to each well. The culture plate was placed into the microplate reader within the specified time. Then the absorbance value of the optical density (OD) 450 nm pore was measured. Each group had 3 sets of auxiliary holes.
Colony formation assay
The density of 2,000 cells was evenly inoculated into a 6-well plate, and after 14 days of culture in the incubator sink, they were fixed with 4% paraformaldehyde for 30 minutes. The cells were then dyed with crystal violet for 20 minutes. After rinsing, photos were taken and the cell colonies were counted.
Flow cytometric analysis
Flow cytometry was used to detect cell apoptosis and cycle. At 48 hours after transfection, double staining with an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) Apoptosis Kit (Invitrogen) was performed, and the cell percentage was stained using a PI-based cell cycle assay kit (Invitrogen).
Transwell assay
After digesting the cells, serum-free culture medium was used to blow them evenly and approximately 10,000 cells were added to the Transwell’s upper chamber. DMEM culture medium was added to the lower chamber. The cells were then incubated for 24 hours. After being fixed with 4% paraformaldehyde for 30 minutes, the cells were dyed with crystal violet for 20 minutes.
WB analysis
After loading the sample, the protein was completely separated by constant current electrophoresis. After the protein was transferred to the polyvinylidene fluoride (PVDF) membrane, it was sealed for 1 hour in sealing solution, and finally the antibody was added to continue incubation overnight. On the second day, after rinsing in tris-buffered saline with Tween 20 (TBST), horseradish peroxidase (HRP)-labeled secondary antibodies were added and incubated at room temperature for 1 hour. Photos were taken with an automatic chemiluminescence imaging system. Primary antibodies were generated against TMEM43 (diluted 1:500, #ab15379, Abcam, Cambridge, UK), VDAC1 (1/5,000, #55259-1-AP, Proteintech, Rosemont, IL, USA), USP7 (1/10,000, #66514-1-Ig, Proteintech), p-PI3K (1/1,000, #AF3242, Affinity, Cincinnati, OH, USA), and PI3K (1/1,000, #MA1-74183, Thermo Fisher, Waltham, MA, USA), and β-actin (1:10,000, #66009-1-Ig, Proteintech) was used as an internal control.
Cellular immunofluorescence
HCC cells were fixed with methanol and washed with phosphate-buffered saline (PBS) for 3 days ×5 minutes. After the first antibody had been added, they were sealed with goat serum overnight. After being washed in PBS, the cells continued to incubate with the second antibody for 2 hours. After cleaning, 4',6-diamidino-2-phenylindole (DAPI) was added and microscopic examination was performed.
Coimmunoprecipitation (co-IP)
The total cell lysate antibody was mixed and incubated overnight. Then, 5 µL agarose (A+G) (Absin, Shanghai, China) was added and incubated at room temperature for a further 2 hours. The composite was rinsed 3 times with PBS. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and WB were used to detect proteins.
Statistical analysis
The software SPSS 26.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis, and the data were calculated as mean ± standard deviation. Independent sample t-test was used to compare the two groups of data, and P<0.05 was considered statistically significant.
Results
Exploring genes regulating HCC through RNA-seq
MiR-7 has been described as an important factor in cancer development as it acts as a tumor suppressor and regulates a large number of genes involved in cancer development and progression (15). To explore potential genes that regulate HCC progression, we first transfected Huh7 cells with empty lentivirus (NC) and miR-7 overexpressed lentivirus (OE), respectively. Detection by qRT-PCR showed a significant increase in expression in the OE group (Figure 1A). Then, we performed RNA-seq on the cells and found that compared to the miR-7 overexpression group (E8321-1, E8321-2, E8321-3), the miR-7 control group (E8320-1, E8320-2, E8320-3) had a total of 342 upregulated genes and 369 downregulated genes. The difference between the two groups was visualized through a volcanic map (Figure 1B).
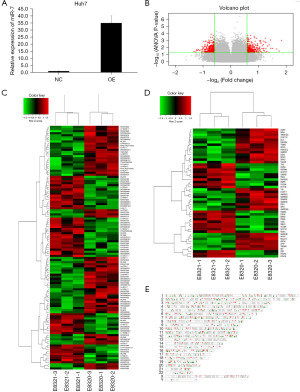
We conducted cluster analysis on long non-coding RNA (lncRNA) and messenger RNA (mRNA) in differentially expressed genes (DEGs) through bioinformatics analysis. We found that compared to the OE group, lncRNA with increased expression in the NC group included lncRNAs ARID1B-2:1, C15orf48-2:1, OTTHUMT000000473305, ENST000000421513.1, ENST000000472821.1, ENST000000624858, and MEF2C-3:1 (Figure 1C). The mRNA with increased expression in the NC group included DPPA3, NPHS1, DGKG, GABRR3, POM121L2, GPR15, TRIM43, FAM183A, DLL1, CCDC110, TMEM132C, NOX4, A4GNT, and TMEM43, among others (Figure 1D). Then, we further analyzed the distribution of DEGs on chromosomes (Figure 1E).
Predicting high expression of TMEM43 gene in HCC
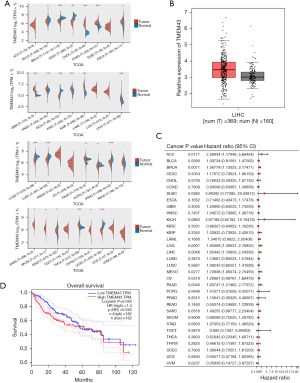
Through RNA-seq, we found that TMEM43 may play an important role in HCC. Therefore, we used TGCA to explore the expression of TMEM43 in 33 cancers. We found that TMEM43 is highly expressed in various tumors, including HCC (Figure 2A). We further confirmed through the Gene Expression Profiling Interactive Analysis (GEPIA) database that TMEM43 is highly expressed in cancer (Figure 2B). Then, we further explored the prognostic impact of TMEM43 in 33 cancers through TGCA and found that TMEM43 affects the prognosis of multiple tumors (Figure 2C). The GEPIA database further confirmed that TMEM43 is closely related to the poor prognosis of HCC (Figure 2D).
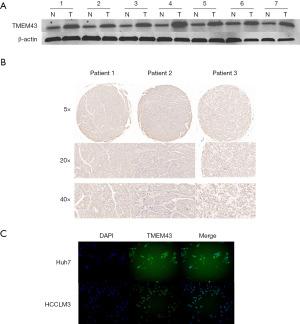
Expression levels of TMEM43 gene in HCC tissues and HCC cell lines
In order to determine the expression of TMEM43 in HCC tissue, we used WB to analyze the expression of TMEM43 in HCC tissue. The results showed that compared with adjacent tissues, the expression of TMEM43 in HCC tissue was significantly increased (Figure 3A). We conducted a tissue microarray study on the expression of TMEM43 in 96 HCC tissues, and found that it was highly expressed in 68 tissues. TMEM43 was mainly brownish brown in the cytoplasm (Figure 3B). Then, we investigated the expression and localization of TMEM43 in Huh7 and HCCLM3 cells through cellular immunofluorescence. The results showed that TMEM43 was mainly expressed in the cytoplasm of HCC cells (Figure 3C).
WB was used to detect the transfection effect of TMEM43 gene in HCC cell lines
To verify the impact of TMTM43 on the biological behavior of HCC cell lines, we used LipofectamineTM 3000 for transfection. We transfected sh-TMEM43#1, sh-TMEM43#2, and sh-TMEM43#3 into the HCC cell lines Huh7 and HCCLM3. WB was used to detect the expression of TMEM43. The results showed that compared with the control group, Huh7 and HCCLM3 cells transfected with sh-TMEM43#1, sh-TMEM43#2, and sh-TMEM43#3 showed significant downregulation of TMEM43 expression. Then, we selected sh-TMEM43#1 and sh-TMEM43#2 with better transfection effects for subsequent experiments (Figure 4A).
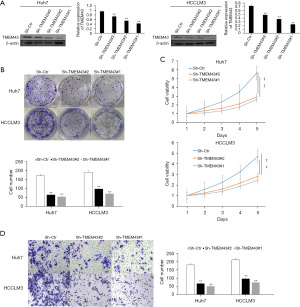
The effect of TMEM43 gene on the growth of HCC cells
The colony formation assay and CCK-8 assay were used to detect the effect of sh-TMEM43 on growth, respectively. The colony formation experiment showed that the cell colony of TMEM43 knockdown group was smaller than that of the control group (Figure 4B). The CCK-8 experiment showed that compared with the control group, TMEM43 knockdown significantly inhibited the proliferation of Huh7 and HCCLM3 cells (Figure 4C). Through CCK-8 and clone formation experiments, it was revealed that knocking down TMEM43 inhibits the growth of HCC cells.
The effect of TMEM43 gene on the invasion of HCC cells
We used Transwell experiments to detect the effect of TMEM43 on the invasion ability of HCC cells. The Transwell experiment results showed that knocking down TMEM43 significantly inhibited the invasion of Huh7 and HCCLM3 cells (Figure 4D).
The effect of TMEM43 gene on cell cycle and apoptosis of HCC cells
Flow cytometry was used to detect the effects of TMEM43 on cell cycle and apoptosis, respectively. Flow cytometry experiments showed that the apoptosis rate of Huh7 and HCCLM3 cells was significantly higher after knocking down TMEM43 compared to the control group (Figure 5A). Flow cytometry experiments showed that compared with the control group, the G1 phase cells of Huh7 and HCCLM3 cells were significantly increased after knocking down TMEM43, whereas the S phase cells were significantly reduced (Figure 5B). Through cell cycle and apoptosis experiments, it was found that knocking down TMEM43 inhibits the growth of HCC cells and promotes cell apoptosis.
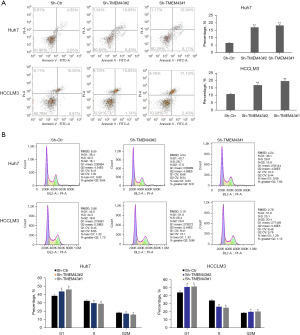
The protein interaction relationship between TMEM43 gene and VDAC1 gene
In Huh7 and HCCLM3 cells, knockdown of TMEM43 resulted in a decrease in the protein level of VDAC1, whereas overexpression of VDAC1 partially restored the knockdown effect of sh-TMEM43 (Figure 6A). Meanwhile, cellular immunofluorescence revealed that TMEM43 and VDAC1 were co localized in the cytoplasm (Figure 6B). The results of co-IP indicated the interaction between TMEM43 and VDAC1 at the protein level (Figure 6C). These results indicate that TMEM43 affects the progression of HCC through VDAC1.
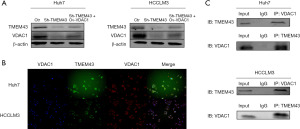
TMEM43 gene affects the proliferation and invasion of HCC cells through VDAC1 gene
To determine whether TMEM43 promotes the proliferation and invasion of Huh7 and HCCLM3 cells through VDAC1, we restored the expression of VDAC1 inhibited by sh-TMEM43 through a recovery experiment. Through cloning (Figure 7A), CCK-8 (Figure 7B), and Transwell experiments (Figure 7C), it was found that overexpression of VDAC1 partially restored the inhibitory effect of sh-TMEM43 on cell proliferation and invasion. The above results indicate that TMEM43 promotes the proliferation and invasion of Huh7 and HCCLM3 cells by targeting VDAC1.
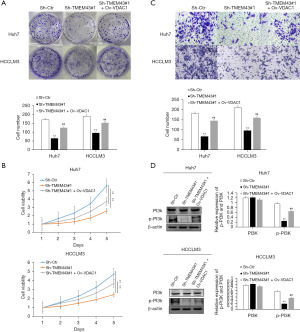
To further investigate the mechanism of TMEM43 inhibiting the proliferation and invasion of HCC cells through VDAC1, we investigated the effect of TMEM43 on the PI3K signaling pathway through WB. We found that sh-TMEM43 inhibited the PI3K signaling pathway in HCC cells, whereas VDAC1 partially restored the inhibitory effect of sh-TMEM43 on HCC cells (Figure 7D). The above results indicate that TMEM43 promotes the proliferation and invasion of Huh7 and HCCLM3 cells by targeting VDAC1.
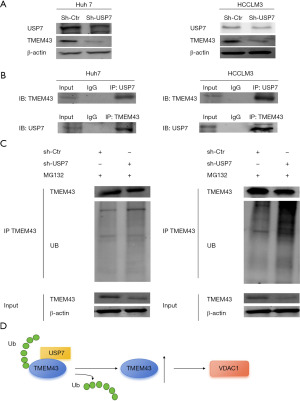
USP7 gene promotes the progression of HCC by regulating the level of TMEM43 gene through deubiquitination
In Huh7 and HCCLM3 cells, USP7 knockdown resulted in a decrease in the protein level of TMEM43 (Figure 8A). The interaction between TMEM43 and USP7 at the protein level was found through immunoprecipitation experiments (Figure 8B). Further, immunoprecipitation revealed that after USP7 knockdown, the level of TMEM43 increased (Figure 8C). The above results indicate that USP7 affects the progression of HCC by regulating the ubiquitination level of TMEM43 through deubiquitination (Figure 8D).
Discussion
In recent years, molecular targeted therapy for HCC has remained a research hotspot (16,17). Therefore, finding effective key therapeutic targets is of great significance for the treatment of HCC. In the occurrence and progression of HCC, multiple oncogenes and tumor suppressor genes all play a role (18,19). Compared with normal control samples and low-grade glioma samples, the expression level of TMEM43 is upregulated in high-grade glioma malignant samples. The higher the expression of TMEM43, the lower the survival rate. In vitro and in vivo experiments showed that knockdown of TMEM43 could inhibit the proliferation and metastasis of glioma (20). However, the role of TMEM43 has not been reported in HCC. In this study, we discovered through the TCGA database that TMEM43 is highly expressed in HCC tissues. TMEM43 is closely related to the prognosis of HCC, and high expression of TMEM43 can lead to poor prognosis. We found that the expression level of TMEM43 in HCC tissue was higher than that in adjacent tissues by WB. Through experiments such as CCK-8, clone formation assay, flow cytometry, and Transwell, we found that overexpression of TMEM43 was associated with HCC cell viability, proliferation ability, migration, and increased apoptosis ability of HCC cells. Therefore, TMEM43 can affect the growth, proliferation, and invasion of HCC cells, thereby affecting the occurrence and development of HCC, which undoubtedly provides new therapeutic targets for HCC treatment.
However, the mechanism of TMEM43 in HCC cells is still unclear. In this study, we found that TMEM43 regulates the proliferation, apoptosis, and invasion of HCC through VDAC1. VDAC protein is the most abundant porogen located in the outer mitochondrial membrane (OMM) of eukaryotic cells. As the gatekeeper of mitochondria, it regulates the entry and exit of metabolites, Ca2+, fatty acid ions, and reactive oxygen species in OMM (21,22). VDAC protein has been identified as three isoforms encoded by three homeotic genes: VDAC1, VDAC2, and VDAC3. VDAC1 has the most abundant expression level among them (23). A study has found that VDAC1 is overexpressed in HCC and can serve as a new diagnostic biomarker. VDAC1 is an independent factor in predicting poor prognosis (24). According to reports, overexpression of VDAC1 is closely related to different types of cancer (25-28). VDAC1 may serve as a new pharmacological target for anticancer therapy (29). This study found that overexpression of VDAC1 can partially counteract the proliferation and invasion of sh-TMEM43 on HCC cells. In addition, we found that overexpression of VDAC1 can partially counteract the inhibitory effect of sh-TMEM43 on the PI3K signaling pathway in HCC cells. These results further confirm the inseparable relationship between VDAC1 and TMEM43 in the occurrence and development of HCC.
Through the action of deubiquitinating enzymes (DUBs), deubiquitination is responsible for removing ubiquitin/polyubiquitin chains from protein substrates and reversing the function of ubiquitination (30,31). The post-translational modification of proteins known as ubiquitination is a key process that has been observed to be dysregulated in several types of cancer, including HCC (32). DUBs play an important role in tumor development by removing ubiquitin from substrate proteins through the deubiquitination process (33). Several families of DUBs exist in the human proteome, including ubiquitin C-terminal hydrolase (UCH), ubiquitin-specific proteases (USPs), ovarian tumour proteases (OTUs) and motif interacting with ubiquitin-containing novel DUB family proteases (MINDYs) (34). The diversity of these DUB families highlights the complexity of deubiquitination and its potential involvement in cancer progression. Not unlike most post-translational modification, ubiquitination is also associated with many cell functions, including cell cycle progression, apoptosis, gene transcription, DNA repair, and signal transduction, which regulate cell development and differentiation and tumorigenesis (35,36). Therefore, components in the ubiquitin pathway have been proposed as potential targets for treatment strategies for diseases and cancer (37,38). USP7 is one of the most abundant USPs and plays a multifaceted role in many cellular events, including carcinogenic pathways (39-42). A study has found that USP7 is a promising target for cancer treatment by regulating the MDM2/MDX-p53 pathway (43). We found that USP7 can affect the expression of TMEM43 in HCC by deubiquitination of TMEM43. This study provides experimental and theoretical basis for studying the development and mechanism of TMEM43 in cancer, as well as for subsequent treatment of cancer.
Conclusions
TMEM43 plays a crucial role in HCC. Further analysis indicates that TMEM43 affects the proliferation, invasion, and apoptosis of HCC cells through VDAC1. To our knowledge, this study is the first to explore the role of TMEM43 in HCC. TMEM43 can serve as a promising biomarker for HCC, laying the foundation for further understanding the mechanism of its occurrence and development. These findings provide a new understanding of the pathology and treatment of HCC, and demonstrate the potential of USP7/TMEM43/VDAC1 axis as a new biomarker for monitoring and treating the disease, laying a solid foundation for the development of targeted therapies for invasive cancer.
Acknowledgments
Funding: This work was supported by grants from Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX21_3110 to N.Z.), the National Science Fund for Distinguished Young Scholars (No. 20925520), and the National Natural Science Foundation of China (Nos. 21235003 and 81172503).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-108/rc
Data Sharing Statement: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-108/dss
Peer Review File: Available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-108/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tgh.amegroups.com/article/view/10.21037/tgh-23-108/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study conformed to the provisions of the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Ethics Committee of the Affiliated Hospital of Nantong University (ID: 2022-L062). Informed consent was provided by all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Roberts LR. Sorafenib in liver cancer--just the beginning. N Engl J Med 2008;359:420-2. [Crossref] [PubMed]
- Anwanwan D, Singh SK, Singh S, et al. Challenges in liver cancer and possible treatment approaches. Biochim Biophys Acta Rev Cancer 2020;1873:188314. [Crossref] [PubMed]
- Gilles H, Garbutt T, Landrum J. Hepatocellular Carcinoma. Crit Care Nurs Clin North Am 2022;34:289-301. [Crossref] [PubMed]
- Liu Z, Liu Y, Zhang W, et al. Deep learning for prediction of hepatocellular carcinoma recurrence after resection or liver transplantation: a discovery and validation study. Hepatol Int 2022;16:577-89. [Crossref] [PubMed]
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76. [Crossref] [PubMed]
- Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev 2023;42:629-52. [Crossref] [PubMed]
- Shinomiya H, Kato H, Kuramoto Y, et al. Aberrant accumulation of TMEM43 accompanied by perturbed transmural gene expression in arrhythmogenic cardiomyopathy. FASEB J 2021;35:e21994. [Crossref] [PubMed]
- Gu Q, Xu F, Orgil BO, et al. Systems genetics analysis defines importance of TMEM43/LUMA for cardiac- and metabolic-related pathways. Physiol Genomics 2022;54:22-35. [Crossref] [PubMed]
- Chen Z, Cao Z, Gui F, et al. TMEM43 Protects against Sepsis-Induced Cardiac Injury via Inhibiting Ferroptosis in Mice. Cells 2022;11:2992. [Crossref] [PubMed]
- Li J, Song Y, Zhang C, et al. TMEM43 promotes pancreatic cancer progression by stabilizing PRPF3 and regulating RAP2B/ERK axis. Cell Mol Biol Lett 2022;27:24. [Crossref] [PubMed]
- Lu T, Xu R, Wang C, et al. Bioinformatics analysis and single-cell RNA sequencing: elucidating the ubiquitination pathways and key enzymes in lung adenocarcinoma. J Thorac Dis 2023;15:3885-907. [Crossref] [PubMed]
- Zhang N, Wang LR, Li DD, et al. Interferon-α Combined With Herbal Compound "Songyou Yin" Effectively Inhibits the Increased Invasiveness and Metastasis by Insufficient Radiofrequency Ablation of Hepatocellular Carcinoma in an Animal Model. Integr Cancer Ther 2018;17:1260-9. [Crossref] [PubMed]
- Li Y, Zhang D, Shi Y, et al. Syntheses and preliminary evaluation of [(18) F]AlF-NOTA-G-TMTP1 for PET imaging of high aggressive hepatocellular carcinoma. Contrast Media Mol Imaging 2016;11:262-71. [Crossref] [PubMed]
- Morales-Martínez M, Vega MI. Role of MicroRNA-7 (MiR-7) in Cancer Physiopathology. Int J Mol Sci 2022;23:9091. [Crossref] [PubMed]
- Llovet JM, Pinyol R, Kelley RK, et al. Molecular pathogenesis and systemic therapies for hepatocellular carcinoma. Nat Cancer 2022;3:386-401. [Crossref] [PubMed]
- Arita J, Ichida A, Nagata R, et al. Conversion surgery after preoperative therapy for advanced hepatocellular carcinoma in the era of molecular targeted therapy and immune checkpoint inhibitors. J Hepatobiliary Pancreat Sci 2022;29:732-40. [Crossref] [PubMed]
- Wang JJ, Lei KF, Han F. Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci 2018;22:3855-64. [PubMed]
- Barzaman K, Karami J, Zarei Z, et al. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol 2020;84:106535. [Crossref] [PubMed]
- Jiang C, Zhu Y, Zhou Z, et al. TMEM43/LUMA is a key signaling component mediating EGFR-induced NF-κB activation and tumor progression. Oncogene 2017;36:2813-23. [Crossref] [PubMed]
- Shoshan-Barmatz V, De S, Meir A. The Mitochondrial Voltage-Dependent Anion Channel 1, Ca(2+) Transport, Apoptosis, and Their Regulation. Front Oncol 2017;7:60. [Crossref] [PubMed]
- Shoshan-Barmatz V, Shteinfer-Kuzmine A, Verma A. VDAC1 at the Intersection of Cell Metabolism, Apoptosis, and Diseases. Biomolecules 2020;10:1485. [Crossref] [PubMed]
- De Pinto V, Reina S, Gupta A, et al. Role of cysteines in mammalian VDAC isoforms' function. Biochim Biophys Acta 2016;1857:1219-27. [Crossref] [PubMed]
- Fang Y, Liu J, Zhang Q, et al. Overexpressed VDAC1 in breast cancer as a novel prognostic biomarker and correlates with immune infiltrates. World J Surg Oncol 2022;20:211. [Crossref] [PubMed]
- Zerbib E, Arif T, Shteinfer-Kuzmine A, et al. VDAC1 Silencing in Cancer Cells Leads to Metabolic Reprogramming That Modulates Tumor Microenvironment. Cancers (Basel) 2021;13:2850. [Crossref] [PubMed]
- Zhuo Z, Lin H, Liang J, et al. Mitophagy-Related Gene Signature for Prediction Prognosis, Immune Scenery, Mutation, and Chemotherapy Response in Pancreatic Cancer. Front Cell Dev Biol 2021;9:802528. [Crossref] [PubMed]
- Li T, Wang N, Li S, et al. ANP32B promotes lung cancer progression by regulating VDAC1. Gene 2023;859:147200. [Crossref] [PubMed]
- Liu Y, Zhang H, Liu Y, et al. Hypoxia-induced GPCPD1 depalmitoylation triggers mitophagy via regulating PRKN-mediated ubiquitination of VDAC1. Autophagy 2023;19:2443-63. [Crossref] [PubMed]
- Magrì A, Reina S, De Pinto V. VDAC1 as Pharmacological Target in Cancer and Neurodegeneration: Focus on Its Role in Apoptosis. Front Chem 2018;6:108. [Crossref] [PubMed]
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 2009;10:550-63. [Crossref] [PubMed]
- Park HB, Baek KH. E3 ligases and deubiquitinating enzymes regulating the MAPK signaling pathway in cancers. Biochim Biophys Acta Rev Cancer 2022;1877:188736. [Crossref] [PubMed]
- Sun T, Liu Z, Yang Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol Cancer 2020;19:146. [Crossref] [PubMed]
- Deng L, Meng T, Chen L, et al. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct Target Ther 2020;5:11. [Crossref] [PubMed]
- Abdul Rehman SA, Kristariyanto YA, Choi SY, et al. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell 2016;63:146-55. [Crossref] [PubMed]
- Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature 2009;458:438-44. [Crossref] [PubMed]
- Li S, Zhang H, Wei X. Roles and Mechanisms of Deubiquitinases (DUBs) in Breast Cancer Progression and Targeted Drug Discovery. Life (Basel) 2021;11:965. [Crossref] [PubMed]
- Harrigan JA, Jacq X, Martin NM, et al. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Discov 2018;17:57-78. [Crossref] [PubMed]
- Wang M, Zhang Z, Li Z, et al. E3 ubiquitin ligases and deubiquitinases in bladder cancer tumorigenesis and implications for immunotherapies. Front Immunol 2023;14:1226057. [Crossref] [PubMed]
- Harakandi C, Nininahazwe L, Xu H, et al. Recent advances on the intervention sites targeting USP7-MDM2-p53 in cancer therapy. Bioorg Chem 2021;116:105273. [Crossref] [PubMed]
- Dai X, Lu L, Deng S, et al. USP7 targeting modulates anti-tumor immune response by reprogramming Tumor-associated Macrophages in Lung Cancer. Theranostics 2020;10:9332-47. [Crossref] [PubMed]
- Park SH, Fong KW, Kim J, et al. Posttranslational regulation of FOXA1 by Polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci Adv 2021;7:eabe2261. [Crossref] [PubMed]
- Korenev G, Yakukhnov S, Druk A, et al. USP7 Inhibitors in Cancer Immunotherapy: Current Status and Perspective. Cancers (Basel) 2022;14:5539. [Crossref] [PubMed]
- Huang YT, Cheng AC, Tang HC, et al. USP7 facilitates SMAD3 autoregulation to repress cancer progression in p53-deficient lung cancer. Cell Death Dis 2021;12:880. [Crossref] [PubMed]
Cite this article as: Zhang N, Wang F, Yang X, Wang Q, Chang R, Zhu L, Feitelson MA, Chen Z. TMEM43 promotes the development of hepatocellular carcinoma by activating VDAC1 through USP7 deubiquitination. Transl Gastroenterol Hepatol 2024;9:9.


