
Evolution of the liver biopsy and its future
Introduction
Since its original description by Ehrlich in 1883, the clinical role of liver biopsy has changed dramatically over the years (1). Initially, it was used for determination of glycogen stores in the livers of diabetic patients while in the ensuing years, its indications have become more diverse and numerous (2). It took a few decades for the liver biopsy to be accepted as a standard medical procedure by the medical community. Only after Menghini described “one-second needle biopsy of the liver” in 1958 did the procedure come into more widespread clinical use (2). While liver biopsies are commonly used to evaluate a wide variety of medical disorders, including neoplasms and post-transplant complications, its use is being impacted by improved clinical diagnosis of disorders, and non-invasive methods for evaluating liver tissue. Nevertheless, liver biopsies continue to provide highly valuable information for clinical and research purposes. This review considers the evolving role of liver biopsies in clinical practice over the decades, and describes technologies that are likely to have a significant impact on how they will be used in the future.
Liver tissue can be obtained for clinical analysis in several ways, including percutaneous, transjugular, laparoscopic, and intra-operative approaches. Each of these have advantages and disadvantages. The percutaneous liver biopsy technique remains the standard practice in most medical centers. Whenever a percutaneous needle biopsy is contraindicated (e.g., presence of significant ascites or coagulopathy), a transjugular needle biopsy is preferred. An important advantage of the transjugular approach is the ability to obtain intrahepatic portal pressures that aid in the diagnosis and management of select groups of patients, particularly those with cirrhosis (3). However, transjugular biopsies often render smaller and fragmented tissue samples, making pathologic assessment somewhat difficult. Percutaneous or transjugular biopsies are used to obtain samples from patients suffering from disorders that involve a diffuse change in the liver (3,4). By contrast, image-guided needle biopsies are necessary for focal diseases/lesions (5,6). Laparoscopic liver biopsy enables the gross features of the liver to be assessed prior to taking the biopsy. During laparoscopy apart from needle core biopsy, a wedge of liver tissue (wedge biopsy) can be obtained. Due to the nature of surgery involved, laparoscopic liver biopsy has added costs and the potential for more complications (7). As such, it is used in select settings at most centers e.g., when laparoscopy is being anyway performed for another indication or the lesion is difficult to approach by other means. Liver biopsies (needle or wedge) can also be obtained intra-operatively whenever liver disease is suspected or suspicious lesions are identified. While needle biopsies provide a core of liver parenchyma, wedge biopsies provide a “wedge” of liver tissue taken from it surface and are much larger. In many instances, an abnormal appearance of the liver during surgery for an unrelated procedure (most often cholecystectomy) is the first indication of an underlying liver disease. Liver biopsies harvested intra-operatively have the added advantage of obtaining tissue samples from grossly visible/suspicious lesions. However, these wedge biopsies are often suboptimal for assessment of liver fibrosis and/or inflammation due to the preponderance of Glissen’s capsule, wider portal tracts in subcapsular area, and frequent (but inconsequential) surgically-induced hepatitis (8). Therefore, needle biopsy should be the technique of choice at laparotomy or be used in addition to a wedge biopsy.
Once obtained, liver tissue needs to be triaged for various special procedures e.g., electron microscopy, microbiological cultures, immunofluorescence studies or flow cytometry, based on the clinical differential diagnosis. Thus, it is very important that critical clinical details (e.g., liver enzymes, history of medications/toxin exposure, viral and autoimmune serology) and a clinical differential diagnosis be communicated to the pathologist so that appropriate tissue triage and special procedures can be undertaken without loss of time.
Histological examination is an essential tool for the evaluation of patients with liver disease, particularly those with persistent unexplained liver function abnormalities. Most pathology labs will routinely perform trichrome, hematoxylin and eosin (H&E), reticulin, iron and D-PAS (periodic acid-Schiff stain after diastase digestion) stains on liver biopsies for clinical evaluation of a liver disorder. Under certain circumstances, special sample preparation and fixation may be required, such as placing the tissue in appropriate culture medium (for suspected infections), processing in metal-free containers (for metal quantification), fixation in glutaraldehyde (for electron microscopy), using frozen tissue (for Oil red O stain for the estimation of microvesicular fat), or simply “RUSH/STAT” processing in critically ill patients (e.g., those with fulminant liver failure or the post-transplant situation) (2). Routine histological evaluation of liver biopsies involves assessment of inflammation, cellular changes involving hepatocytes and bile ducts, abnormal accumulation of various substances, the extent of the liver injury, and/or fibrosis. The liver conditions in which a biopsy is most commonly performed include viral hepatitis, non-alcoholic fatty liver disorder (NAFLD), autoimmune disorders, drug/toxin-induced liver injury (including alcoholic liver disease, ALD), cholestatic diseases, metabolic/inherited disorders, and hepatic masses (2,3,5,9-14). In the post-transplant setting, liver biopsies are used for the evaluation of organ rejection, infections, surgical complications, and disease recurrence. However, the indications for liver biopsy in all of these conditions continues to evolve as other approaches to monitoring liver health are developed or diseases that previously necessitated liver biopsies are more effectively treated (2).
Historically, liver biopsies have been used in hepatitis B and C because it permitted evaluation of the stage and severity of the disease, its response to therapy (15,16). Due to high specificity, their less invasiveness, faster turn-around and cost, serological diagnostic tests are now used rather than liver biopsies to establish the diagnosis of hepatitis B or C infection (13,14). A biopsy may be used as an adjunct to serological tests for the assessment of the degree of liver injury and fibrosis, presence of other concomitant liver disease or, under some circumstances, the response to antiviral therapy. The efficacy of direct-acting antiviral (DAA) therapies for HCV infection (>90% cure) make liver biopsies redundant in this condition. Thus, it is not surprising that the use of liver biopsies in clinical practice is declining worldwide.
Another major liver disorder in clinical practice is NAFLD. With better control of viral hepatitis, it is expected that NAFLD will likely become the most common liver disease worldwide. The need for a liver biopsy in this setting is limited as existing imaging techniques are excellent for the evaluation of fat in the liver. However, current methodologies are sub-optimal for assessing inflammations and fibrosis. In the presence of other concomitant liver diseases, the relative contribution from each disease also can be difficult to evaluate without a biopsy. On the other hand, given the limitations of treatment options and ability to predict the disease course based on histology, the biopsies are performed only in select settings in NAFLD. Further, advances in the non-invasive methods (serologic or imaging based) for assessing fibrosis in the liver make the need to liver biopsy even less (13).
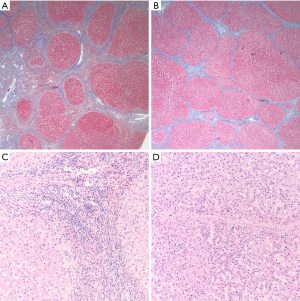
While a variety of histologic changes are evaluated in a liver biopsy for medical disorders, one of the most important parameters is fibrosis which can be indicative of disease progression or response to any treatment. Once a controversial topic, it is now well-accepted that fibrosis can regress, and even cirrhosis can regress (17). In 1979, Pérez-Tamayo first provided evidence in both animal models and human disease showing reversal of fibrosis and cirrhosis (18). Subsequently, serial biopsies from a patient with hepatitis B following antiviral treatment showed regression of fibrosis from fully developed cirrhosis to incomplete septal cirrhosis. In this landmark paper based on livers removed at transplantation having cirrhosis or incomplete septal cirrhosis, Wanless and colleagues reported histologic parameters reflective of progression or regression of fibrosis (19). Subsequent studies reported biopsy-documented regression of cirrhosis associated with hepatitis C, hepatitis B and autoimmune hepatitis following disease-specific therapy (20). Given that liver fibrosis may be reversed by therapeutic intervention, a renewed focus has been placed on tools to monitor fibrosis (and therefore therapy effectiveness). However, assessment of changes in the severity of fibrosis remains challenging. Most investigations have employed fibrosis staging systems that have been in use for several decades; these works well when there is a definite numerical change in the fibrosis stage despite issues of sampling variability. Most common fibrosis scoring systems, such as the Ishak, Batts and Ludwig or METAVIR staging systems, were developed before the idea of fibrosis regression gained importance and, as such, are not equipped for assessing this aspect of fibrosis (21,22). In essence, these systems can detect a change as progression or regression only when there is definite numerical change in fibrosis stage assignment. However, they are not reliable for evaluating changes within a given stage, especially cirrhosis. Historically, cirrhosis has been considered to be “end-stage”. It is now recognized that cirrhosis is a spectrum that varies from mild to severe disease (23). Several systems have been developed to sub-classify cirrhosis, but even these systems fail to predict the course of fibrosis based on a given single liver biopsy obtained at a given point of time (24,25). However, recent studies focusing on fibrosis regression have shown a variety of histopathologic changes that at any given time point suggest fibrosis regression without requiring sequential biopsies (19). Cirrhosis regression is associated with a net decrease in the amount of fibrous tissue, thinning or loss of fibrous septa, disappearance of shunting neo-vessels (that develop as a result of angiogenesis associated with fibrogenesis), disappearance of ductular proliferation and an overall decrease in the cellularity of the fibrous septa (26). Regression of fibrosis is associated with a partial or full restoration of the lobular organization (27). These various features have been taken into account to develop a novel “progressive-indeterminate-regressive (or PIR) system” to assess the dynamic change in fibrosis that can be applied to routine liver biopsies (Figure 1A,B,C,D). This system takes several histologic characteristics into account and provides insight into the quality or directionality of fibrosis—progressive or regressive—in patients with cirrhosis based on a single liver biopsy. This system can be easily applied on H&E and trichome stained slides obtained from formalin-fixed paraffin-embedded tissue. Importantly, it has already been shown to correlate with liver-related morbidity and mortality in the setting of treated chronic hepatitis B (28,29). As this and/or similar systems are validated for routine clinical practice, the evaluation and reporting of liver biopsies will have to adapt to incorporate this aspect of fibrosis assessment.
Thus, over the years, the role of the liver biopsy has been changing from a diagnostic modality to a prognostic evaluation. It is anticipated that liver biopsy will continue to be used in: (I) cases of viral hepatitis with unusual presentation; (II) patients with more than one concomitant disorder; (III) liver disease with unclear etiology; and (IV) liver diseases in which the results of various non-invasive measurements are discrepant with each other or inconsistent with clinical findings. However, the use of liver biopsy in the future is likely to be eroded as new technologies are developed that are less invasive, and able to identify changes in liver structure or function with higher resolution/fidelity.
It is also important to realize that technologies are evolving that allow visualization of liver tissue in three-dimensions, digital morphometry, and analysis of the content of liver cells that go far beyond what immunohistochemistry or other current molecular techniques can accomplish. The application of artificial intelligence (AI) and machine learning (ML) further enhances these technologies by automating their analysis and interpretation.
Role of digital pathology
The application of digital technology has had a significant impact on medicine and pathology has been at the forefront of this ‘digital revolution’. Analysis of images obtained from select parts of histologic tissue sections has been in practice for decades. Only recently development of advanced slide scanners has allowed creation of “digital/virtual” slides that have image of the entire tissue section present in a slide [whole slide imaging (WSI)]. This can now be done very rapidly and at very high resolution, allowing its use even in routine clinical practice for making diagnoses and evaluating prognostic biomarkers. Digitally-scanned tissue sections also permit rapid access and sharing of image data. In addition, it permits the development and application of novel, computer-based methods for analyzing tissue parameters. To this end, AI is being leveraged to automate tissue analyses and enhance processes and accuracy in pathological diagnoses (30). One notable example of this new approach is the increased accuracy and efficiency of AI-assisted diagnosis of lymph node metastasis detection compared to human pathologists (31).
Application of digital pathology to the liver has overwhelming been focused on establishing quantitative measurements of fibrosis. Routine human-based histologic examination of fibrosis has been notoriously fraught with inter- and intra-observer variability and inter-laboratory staining variability. Digital image analysis/morphometry (DIA) techniques increase the level of objectivity and reproducibility of these analyses. While DIA can overcome some of the limitations of tissue artifacts or staining variability, it cannot address the issue of sampling error, as this occurs before the tissue is received in the pathology lab. Variations of this technique have been used for more than thirty years now, and have been shown to correlate with previously established histology-based semi-quantitative measures of fibrosis, such as the Ishak, Metavir, Knodell or Scheuer systems (32-36). In the setting of cirrhosis, DIA techniques correlate with sub-classification staging indices, such as the Laennec or Jain-Garcia systems (37). They have also been shown to correlate well with certain clinical parameters, such as the Hepatic Venous Pressure Gradient (33), and with the progression or regression of fibrosis in the setting of treated chronic viral hepatitis (33,38,39).
Other areas that lend themselves to quantitation by DIA are assessment of steatosis and inflammation (40,41). The degree of fat in liver tissues has been associated with worse prognosis and the development of steatohepatitis in NAFLD (42). In addition, fat in the liver has been associated with worse graft outcomes (43). These represent a few of many reasons justifying comparison of digital morphometric methods for assessing the degree of hepatic fat content by more objective methods (41,44-46). DIA algorithms have recently been designed to be incorporated in large scale research studies, including clinical trials for NAFLD (47).
Despite the advantages and potential of WSI, its adoption in clinical practice has been slow. Slide scanners are very expensive and storing of large quantities of digital data remains an issue. In addition, scanners require trained technicians to operate, and good informatics support is required on the back end. Thus, a major barrier to adoption of WSI has been the associated increase in cost and labor to the already labor-intensive and slow process of standard tissue formalin fixation, paraffin wax-embedding, slice cutting and staining. A second barrier has been the reduction in image quality compared to the currently used transmitted light microscopy (48-50). Although artifacts from focusing error in WSI continue to be reduced, WSI systems will remain limited by artifacts, such as tissue tears, folds and stain variability that are inherent in the process of physically cutting thin tissue slices. None the less, the slide scanners are getting cheaper and more versatile in terms of their speed and DIA applications. Storage capacity of digital data is improving. With these advances, it is expected that the use of DIA will continue to expand as new algorithms are developed that allow objective assessment of variety of other histologic features in liver biopsies, besides fibrosis and steatosis.
Evolving technologies for histologic evaluation of liver tissues
In vivo multi-photon and 3-dimensional microscopy
For over a century, diagnostic histology has relied on thinly cut tissue slices mounted on slides, stained with dyes (most commonly H&E), and examined in 2-dimensions using an ocular microscope using white light. A variety of technologies developed in recent years use different light sources and/or microscopes that allow visualization of tissues in a completely different way. For example, multiphoton microscopy (MPM) (described in more detail below) allows visualization of tissues in 3-dimensions based on fluorescence excitation and detection (51-53). It has been applied widely in research and is being developed for clinical use. Other exciting advances such as new techniques for improvement of deep imaging of tissue via chemical clearing and advanced image analysis methods such as convolutional neural networks (CNNs) (see below), when combined with MPM microscopy, portend dramatic improvements in the efficiency and precision of histologic evaluation of liver tissues.
Advances in optics over the past few decades have generated an array of potential alternatives to formalin-fixed, paraffin-embedded tissue (FFPE) and WSI for digital image analysis of specimens without thin physical sectioning or wax-embedding, including liver. The approaches can generally be categorized into the following: optical coherence tomography (OCT) (54), stimulated Raman spectroscopy (SRS) (41,55), selective plane illumination (light-sheet) microscopy (SPIM) (56), deep UV surface imaging [e.g., microscopy with ultraviolet surface excitation (MUSE)] (57), confocal microscopy (58), and MPM (59,60). Other modalities exist but generally involve higher complexity, making their near-term applicability to routine clinical use less likely.
It is important to appreciate that all of these new methods require less processing than the standard histological tissue preparation, and they are intrinsically digital. Several require very little to no tissue processing time (i.e., OCT, SRS, MUSE), of particular interest for time-sensitive applications such as intraoperative evaluation. Others enable three-dimensional visualization (i.e., SPIM, confocal, MPM), an approach of still theoretical interest for clinical evaluation of tissue samples. All of these approaches avoid tissue cutting artifacts and generate morphologic information without consuming valuable tissue. As such, they improve the yield of informative data that is extractable from a given sample relative to the commonly-used physical slices.
Despite these benefits, these alternative imaging techniques, often grouped under the misnomer of ‘ex vivo’ imaging, face challenges of their own for clinical adoption. Slow imaging time is a critical limitation of several of these approaches (especially SRS and the standard implementations of confocal and MPM) which may take many minutes or hours to generate a single biopsy optical slice, and thus are unable to achieve adequate throughput for clinical use. Some (e.g., SPIM) are limited by the computationally-intensive image processing. This may be addressable by more computing power, but this adds to analysis cost and complexity. The perpendicular optical setup of SPIM also makes specimen orientation incompatible with clinical workflow in its standard configuration, for which specialized approaches have been devised that permit placing a specimen on a flat surface and imaging at an angle (61). Finally, those methods that produce data that are not directly convertible to images using standard histologic stains, e.g., H&E, require pathologist retraining; this represents an additional barrier to adoption. The overarching limitation of these so-called ‘ex vivo’ imaging approaches is that all (except the confocal and MPM techniques) produce images that are of inferior quality relative to standard manual transmitted microscopy. As such, they should only be considered as an adjunct to existing methods for definitive diagnosis. Given the irreversible and often more marked distortion of morphology associated with the use of frozen sections, several of these new approaches may offer a viable alternative for intraoperative evaluation, even in their current state of development. Nevertheless, only those methods with the highest quality imaging, such as confocal and MPM, are likely to be able to move histology beyond standard physical slide use.
The image quality delivered by many of these new approaches can be increased by incorporating another recent advance in imaging called “clearing”. This involves making a tissue transparent through chemical treatment. A clearing step has been a part of standard histology processing for nearly a century, and it referrs to the partially cleared appearance that the use of xylene imparts in its role as an intermediary reagent with solubility in both alcohol and molten paraffin wax. With the advent of confocal microscopy, MPM, and SPIM for deep imaging of uncut, un-embedded tissue specimens, there has been a resurgent interest in the development and use of chemicals that can match the optical properties of proteins and membranes (i.e., the high refractive index of these components), thereby improving the quality and extending considerably the depth within tissue at which fluorescent images can be obtained. The past few decades have engendered a host of approaches and chemical compositions for improving clearing of tissues which allows better light penetration (62). The approaches can be divided into those that involve (I) electrophoretic removal of lipids (e.g., CLARITY and its extensions), or (II) the replacement of the water with a high refractive index fluid, e.g., BABB, 3DISCO, etc. While of potential value for investigative use due to high transparency, which improves light collection at depth, electrophoretic removal of lipids presents particular challenges for clinical use in that it is more labor intensive and more complex than pure chemical clearing. By removing membrane and membrane bound molecules, it may also limit the reliability of certain immunohistochemical (IHC) stains in routine clinical use, making it incompatible with the current diagnostic standard. On the other hand, chemical clearing-is a processing step that can allow fluorescence microscopy to achieve contrast, clarity, and resolution surpassing that of transmitted light microscopy. This offers an opportunity for improvement in the reliability of diagnostic interpretation of liver.
The quality of digital images ties into the emerging advances in computerized image analysis. Just as pathologists require quality images for interpretation, computerized classification depends on high quality image data. The introduction of CNNs has significantly advanced automated image pattern recognition. The method involves mathematically combining (convolving) known patterns, such as repeating black and white columns adopting varying frequencies and orientations, with portions of an image. The resulting nodes are then convolved further with other patterns to form layers of a network. Deep networks refer to algorithms that involve many layers, each progressively representing higher order patterns and from which the training process can select the most relevant to a given histologic feature recognition, biological process, or cell type. In the past few years, CNN methods have begun to perform as well as humans at image pattern recognition for specific tasks. Given this rapid progress, it is expected that such digital image interpreting tools will aid pathologists in reproducibly grading and accurately recognize disorders. In as much as they provide higher quantity and potentially higher quality of digital data, techniques such as MPM and confocal can be combined with the new image analysis developments to augment the efficiency and precision of microscopic diagnostic assessment further.
New imaging: applications to liver disease
Owing perhaps to the use of commercial confocal and multiphoton-based in vivo endoscopes for diagnostic liver assessment, these modalities have seen the greatest use in ex vivo liver samples (58,63,64). In unstained liver specimens, confocal microscopy and MPM allow visualization of overall cellular features using intrinsic fluorescence derived primarily from nicotinamide adenine dinucleotide hydride (NADH), its phosphorylated form (NADPH), and flavin adenine dinucleotide (FAD) (59,64,65). Such unstained approaches have been applied to the identification and characterization of liver cancer and steatosis using standard H&E stained sections as the reference (41,66).
Added information can be derived by exploiting the finding that the typical length of time a fluorophore spends in an excited state before releasing its light energy (known as the fluorescence lifetime) varies depending on the fluorophore and its microenvironment. Specialized detection systems can be combined with MPM excitation in a modality called fluorescence lifetime imaging (FLIM) to produce image patterns with enhanced biological information, such as recognition of certain cell types, reduction-oxidation state, and apoptosis/necrosis (67). Unfortunately, this detection process requires more extended imaging times and expensive equipment, reducing its clinical potential for cost-effective routine analysis.
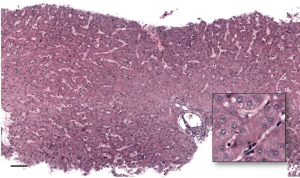
A somewhat more established role for MPM in liver analysis is measurement of collagen fibrosis (53,60,68-70). Although multiphoton laser excitation systems are more expensive than confocal laser systems, MPM has several advantages over confocal for clinical (and investigative) liver imaging. Specifically, it (I) can image deeper into tissue (typically ≈200 µm, depending on fixation and clearing state), (II) has lower risk of damaging tissue or nucleic acids from high intensity light, and (III) can access the collagen-specific signal derived from second harmonic generation (SHG) (Figure 2). Collagen is nearly the only liver molecule that, due to its non-centrosymmetric properties, can combine two photons arriving nearly simultaneously into one photon of exactly twice the energy. This SHG signal differs from fluorescence signals and occurs at exactly half the wavelength of the excitation wavelength so it can be readily separated during imaging, producing a digital map of collagen fibrosis.

SHG produces a relatively reproducible digital collagen distribution signal. As such, it has been touted as a potential means to improve upon the recognized low concordance of fibrotic scoring by visual estimation, which occurs even when collagen specific stains, such as Masson’s, Picrosirius Red, and van Gieson, are used (71). Underscoring this contention, a commercial instrument has been developed that uses SHG derived via MPM on unstained slides for liver fibrosis assessment (71). It also employs algorithms analyzing fiber distribution for additional fibrosis characterization. It is important to appreciate, however, that SHG is relatively specific for the detection of type I and II collagen. Given that Masson’s stain detects type I and large type III collagen fibers, and Picrosirius Red detects both type I and III fibers, SHG patterns may differ from these standard stains. Nevertheless, overall studies have shown good correlation. Still, when using physical slides, SHG does not save on labor, adds to cost and time, and it is unclear whether the analysis has clinical significance beyond image analysis assessment that can be derived from WSI of standard collagen stains, a process that does not require a specialized instrument.
MPM, clearing, and CNNs in liver disease
Implementation of new histological techniques in clinical practice is most likely to succeed only if it can satisfy several key requirements: (I) it must be fast enough to be compatible with the expected clinical volume; (II) it must reduce (rather than increase) labor requirements; (III) it should not create an undue burden at other steps of the overall process (such as surgical collection); (IV) it must improve diagnostic yield; and (V) it must be cost-effective to perform. MPM has been combined with advanced clearing and fluorescent stains that mimic H&E in an approach that satisfies the requirements for practical clinical implementation. Although it has not yet been extensively validated, the technique known as Clearing Histology with MultiPhoton Microscopy (CHiMP) is also applicable to liver specimens and is capable of producing images that look like standard H&E preparation that may be used for primary diagnostic assessment (Figure 3) (60,72). It has the potential to decrease inefficiency, reduce artifacts, and increase data while being readily integrable into clinical workflows.
Using a prototype specialized MPM with high speed acquisition, CHiMP can image with sufficient speed to adequately match clinical volumes, taking ≈1 minute per optical slice. Multiple levels can be evaluated remotely within hours of biopsy. Since no tissue is consumed, specimens may be subsequently processed for standard histologic stains or nucleic acid analysis, as dictated by the morphology. A sample preparation method developed for detailed 3-dimensional collagen fibrosis assessment in liver samples in the research environment (52) could be extended to clinical samples. The additional clinical validation required would be worthwhile because the technique offers considerable improvements over the current histology standard and the real possibility of clinical use in the near future. Like other ‘ex vivo’ imaging modalities, a feature of CHiMP that is particularly significant for its application in diagnostic liver pathology is that it is intrinsically digital and devoid of the artifacts created by physical slicing and standard transmitted light microscopy and WSI. This means clearer images for remote interpretation by expert liver pathologists, and improved data for computerized image analysis. In addition, MPM can produce very thin optical slices, and thereby provide clarity beyond what is achieved with routine histologic sections. As a result, it is anticipated that the data generated by CHiMP will be particularly well suited for use with ML algorithms, including CNNs.
CNNs and other image analysis tools have been applied to H&E slides (73,74), collagen stains (75), and, less commonly, MPM of liver tissue for the identification of clinically-relevant findings (76). Published results thus far show considerable promise for efficient, reproducible, and quantitative assessment for a variety of tasks, such as assessment of lymphocytic inflammation and ploidy. The use of CNN has engendered tools that are already being offered for clinical use in radiology. Unfortunately, the story is different for histology. This is, in part, due to the fact that microscopic slide data are an order of magnitude more complex than radiologic images. In addition, there is considerable variability stemming from stain variability, cutting artifacts, and WSI system imagers. As a result, general applicability is a challenge for CNNs. In this regard, high quality MPM images (such as those derived using CHiMP) could enable reliable clinical implementation. There is a pressing need for studies that can demonstrate the utility of CNNs as clinical diagnostic aids in MPM images of liver disease. Along with operationalized cost-effective instrumentation, these data could stimulate rapid adoption of these new imaging techniques as a new standard for microscopic liver assessment.
Radiologic imaging techniques have demonstrated significant improvements in diagnostic precision that have reduced the clinical need for standard histologic analysis. Given that ‘ex vivo’ tissue can be manipulated in ways that yield far more detailed morphologic and compositional information, biopsies will continue to be able to provide diagnostic data beyond what in vivo analysis can yield. Together with the techniques described in the section that follows, the new microscopic imaging modalities, particularly those based on MPM coupled with the refinement of rapidly evolving ML approaches, are poised to usher in a new era of liver disease diagnostics.
Role of matrix assisted laser desorption/ionization (MALDI) in evaluation of liver tissues: successes, challenges, and future development
Clinical application of MALDI
Obtaining a liver biopsy is an invasive procedure. Consequently, it is of paramount importance that the tissue sample yield clinically-relevant information that cannot be obtained by non-invasive means. MALDI mass spectrometry is a technology that can provide rapid and in-depth characterization of thousands of molecules in a tissue sample. Briefly, laser pulses are directed onto the surface of a tissue coated with a crystalline matrix that is used to extract biological molecules from the tissue. The laser pulses cause ions to be emitted from both the matrix and biological molecules; the ions enter a mass spectrometer where the entire mass range of molecules of interest can be measured (mass spectra) or the ions can be further fragmented for structural confirmation. Therefore, from a single point on a liver tissue sample (usually in the range of 30 to 500 µm), hundreds to thousands of molecules can be identified.
From a clinical perspective, this approach can be applied in two manners (Figure 4). First, MALDI can be used for high throughput screening (77,78). High throughput screening is accomplished by rapidly obtaining measurements of biological molecules across a large number of samples via selecting specific tissue regions, placing cell cultures in well plates, and/or increasing the diameter of the laser. Second, in a more time intensive experiment (hours to days), the approach can be used to generate a molecular image. While the time to obtain MALDI images may be prohibitive in the context of diagnostics, except for cases where no other sufficient tool exists, MALDI imaging can be used to determine a region of interest where diagnostic markers may be found, and then a higher throughput approach as described above can be developed.
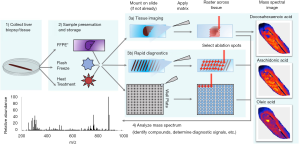
A molecular image in the case of MALDI, is an image showing the spatial distribution of individual molecules (for example proteins, peptides or metabolites) at micrometer resolution. The image is obtained by rastering the laser across a tissue region, generating mass spectra across the tissue at every laser pulse. The resulting data contains “molecular images” often for hundreds to thousands of molecules: the clinician or researcher can use software to select molecules of interest and see the distribution of numerous molecules from a single MALDI analysis. Depending on the setup of the mass spectrometer and choice of matrix, MALDI mass spectrometry can be designed to investigate a diverse range of compounds, including metabolites, lipids, proteins, peptides, and exogenous molecules, e.g., drugs (79).
Applications of MALDI to liver biopsies
As discussed earlier, non-invasive procedures are replacing liver biopsies for assessment of a variety of parameters; however, liver biopsies continue to play an essential role in clinical practice. In all cases, it is advantageous to have in-depth, spatially-resolved molecular measurements that can both describe the current condition of the liver and predict outcomes based on a detailed understanding of liver molecular biology. In these regards, MALDI has been applied in a clinical research setting with promising diagnostic and prognostic capabilities (including differentiating disease states that closely resemble each other and differentiating stages of the disease), as well as being able to elucidate mechanisms of liver diseases that may be targeted for the development of new therapies. Clinically-relevant applications of MALDI include gastric cancer spread to the liver (80), liver metastasis of pancreatic cancer (81), liver cancer (82), colorectal cancer (83), liver infection (84,85), and non-alcoholic fatty liver disease (86). Its application in other areas of liver diseases, such as toxin- or drug-induced injury, metabolic disorders, or infiltrative disorders (e.g., amyloid), have yet to be explored.
The most promising and well-studied areas for the application of MALDI to liver disease are currently liver cancer and NAFLD. Liver cancer is expected to be the 3rd leading cause of cancer-related mortality in 2030, and its prevalence is increasing at a rate of 3.7% in men and 2.9% in women annually (87). Despite extensive research, hepatocellular carcinoma (HCC) remains a cancer with poor prognosis and very limited treatment options. Serum alpha-fetoprotein is the only clinically-used tumor biomarker for screening and surveillance of HCC. However, it suffers from low specificity and sensitivity (88). Surgical resection or transplantation are the only curative options, but the outcomes are highly variable (88). In patients with an established diagnosis of HCC, the need for better tools for prognosis, distinguishing it from similar cancers, and assessment of margins during surgical resection cannot be overemphasized.
Currently, the assessment of surgical margins for HCC resections is based on histological examination of the tissues (89). Even with the use of this methodology, recurrence rates remain high and prognosis is still very poor (89). It is anticipated that more refined assessments of margins based on molecular signatures of involved and uninvolved tissues could lead to better outcomes. In this context, MALDI technology has been applied to differentiate tumor from non-tumor tissues at a molecular level, and it appears to be very promising. Han and colleagues conducted MALDI imaging of peptides in flash frozen liver biopsies to better understand the disease progression and to identify potential targets for treatment (89). Using their classification models, the authors were able to distinguish tumor from non-tumor tissues and to study the tumor borders. Unfortunately, they did not identify the specific peptides distinguishing these tissues. Another factor important in the prognosis of patients with HCC is microvascular invasion. Its assessment by routine pathologic evaluation is fraught with many issues, including sampling and subjectivity in interpretation (90). Using MALDI imaging, Poté and coworkers determined modified forms of histone H4 that could be used to identify regions of microvascular invasion. Importantly, they were able to validate their findings in a second cohort of patients (91).
An additional factor important in prognosis and the choice of treatment is the accurate diagnosis of HCC in relation to the stage of progression and its distinction from other similar neoplasms or healthy adjacent liver. MALDI has been applied to liver biopsies to distinguish (I) HCC from cirrhotic nodules using monomeric ubiquitin in cancerous regions (92), (II) hilar cholangiocarcinoma from peripheral cholangiocarcinoma using neutrophil peptides (93), (III) fibrotic liver from HCC using classification models of unknown peptides (94), and (IV) HCC from metastatic colon cancer using protein-based classification models (95). MALDI has also been used to study progression of HCC using the ratio of lysophosphatidylcholine to phosphatidylcholine (96). In this latter study, the enzyme lysophosphatidylcholine acyltransferase 1 (LPCAT1) was shown to modulate phospholipid composition such that it favored HCC growth. This led LPCAT1 to be suggested as a potential drug target for HCC treatment (96). These studies illustrate the capacity of MALDI to identify a single molecule (or molecular signature) that is a sensitive and specific diagnostic or prognostic marker. In addition, MALDI promotes a greater understanding of the molecular mechanisms involved in the development of HCC and other liver diseases.
The incidence of fatty liver disease (and associated cirrhosis) is increasing globally (97). In the United States, it has been associated with increasing incidence of HCC, and has become the leading cause of liver disease-related mortality (97). Two of the most common fatty liver diseases are alcoholic fatty liver disease (AFLD) and NAFLD. Both are characterized by steatosis of the liver, which can be complicated by steatohepatitis, progressive fibrosis, cirrhosis and/or HCC. While NAFLD and AFLD have similar mortality rates, the underlying cause of mortality and associated risk factors differ significantly (98). It is clinically very important to differentiate between these two diseases because the clinical management and options for transplants differ (98). However, such differentiation is not always easy, and sometimes these diseases may coexist in the same patient (99). In clinical practice, liver biopsy and/or various non-invasive imaging techniques (including ultrasonography, computed tomography, and magnetic resonance imaging) are used to diagnose fatty liver disease, identify the cause, and evaluate disease progression (100,101). However, these techniques have significant limitations for differentiating NAFLD from AFLD in the clinical setting (101).
Lipids play a key role in fatty liver disease pathology (86,102-104). As such, it is anticipated that lipid profiles are likely to be different based on the etiology. Since MALDI allows rapid high-throughput analysis of tissue samples for lipids, it could be a very useful approach for differentiating the different etiologies of NAFLD and AFLD. Lipids can be directly analyzed by MALDI. In AFLD, ethanol increases lipogenesis by up-regulation of sterol regulatory element binding protein 1 (105). In addition, ethanol promotes the formation of lipid droplets in the liver, and reduces the secretion of very-low-density lipoprotein (VLDL) (105). Other important lipid changes associated with fatty liver disease are those involved in inflammatory responses, e.g., an increase in sphingolipids (106). There are also more direct changes related to cell damage, e.g., generation of oxidized lipid species (107). Following binge drinking, ethanol levels in the liver can attain high millimolar concentrations (108), and lead to the generation of lipid-ethanol adducts, specifically phosphatidylethanol species. The presence (and levels) of these lipid molecules represent a potential indicator of harmful and prolonged exposure to ethanol (109), especially given that many of them are only produced in the presence of ethanol (110). MALDI, which can identify thousands of lipids at low micrometer resolution in a tissue section, thus has great promise for elucidating subtle tissue region-dependent differences. This can be used to identify specific changes in the liver during disease development and progression to an extent that is unrivalled by any conventional approaches. MALDI has been applied previously to NAFLD with great success to determine lipid heterogeneity, with the distinct distribution of lipids in NAFLD aiding researchers understanding of disease pathology and its classification (86,102-104). Using imaging mass spectrometry, various lipid molecules have been shown to localize to specific regions of the liver and to correlate with progression from normal to NAFLD to non-alcoholic steatohepatitis (NASH) (102). For example, lipidomics, the comprehensive measurement of lipids within a biological substrate or system, was able to distinguish NAFLD from NASH using specific glycerophospholipids, sphingolipids, and sterols (111). MALDI analysis of lipids has been used to assess disease progression in NAFLD, showing loss of lipid zonation with disease progression (102). The ability to measure the density and size of steatotic regions using specific lipid species [including certain phosphatidylglycerol (PG) species: PG(18:1_20:4) and PG(18:2_22:6)] also helps in this regard (86).
Another potential application for MALDI in the clinical setting is the diagnosis of infections. The liver can be affected by a variety of infections, the most common being viral hepatitis A, B and C (112). While diagnosis of these common infections in clinical practice is seldom a problem, detection of other infections can be very expensive and challenging. Each of these infections can contribute to the development of acute liver failure, chronic hepatitis, fibrosis, and cirrhosis (112). The application of MALDI in the clinical setting for detecting pathogenic microbes and differentiating related species (including viruses) has been shown (85). While generally applied to microbial cultures from blood, the technique could readily be used in liver tissues. As currently used, cell cultures from patients are placed into MALDI wells, treated with formic acid, and a matrix is applied. Common MALDI well plates contain 98 wells, and once cultures are added to wells the instrument can rapidly obtain spectra moving from well to well and compare these spectra against libraries of spectra unique to various organisms for identification.
MALDI has been effectively applied in the clinical laboratory to identify various aerobic (113), gram-positive (114,115) and gram-negative bacteria (116), and yeasts (117). Extension of such approaches to liver tissue for the diagnosis of hepatic infections in the clinical setting may be a more accurate, comprehensive, and cost-effective tool as compared to conventional approaches.
Due to the capacity to identify and measure specific proteins and lipids (and their derivatives) in specific regions of liver tissue, MALDI has great potential for the identification of specific protein deposition disease (e.g., amyloidosis), drug-induced liver injury, and metabolic liver disease. The application of MALDI in these areas for potential clinical use warrants further exploration.
Is MALDI ready for the clinical laboratory? Successes, limitations, and future directions
There have been a number of promising diagnostic and prognostic omics-based tools that have been explored in the last decade; only a few have made it into the clinic. This is for a number of reasons that include: (I) no follow-up validation studies or failure during validation; (II) high costs of analysis and need for highly trained personnel; (III) low reproducibility; and/or (IV) rejection by regulatory agencies or the medical community due to lack of familiarity (118-120). Early research using MALDI has shown tremendous potential for application to liver tissues for diagnosis and prognosis.
However, cost represents a significant factor that could limit the adoption of MALDI in the clinical setting. The instrument alone can approach a million dollars (depending on the desired performance and service contracts), and personnel training adds to this cost. Surprisingly, the few studies that have done a cost analysis of applying MALDI in the clinical setting showed that its implementation for identification of microbes in cell cultures of patients with infections not only improved patient outcomes but also led to cost savings. For example, a study from the University of Michigan Health System showed that implementing MALDI for rapid diagnosis of blood stream infections decreased thirty-day mortality compared to traditional methods (12% vs. 21%, respectively), and total hospital costs decreased per patient by ~5%, saving the hospital ~$2 million dollars annually (118). Of the studies conducted thus far, this is the most thorough cost-effective analysis of MALDI implementation in a clinical setting. That being said, a few other studies have also showed financial benefits of implementing MALDI. A study from the University of North Carolina showed that the savings due to application of MALDI for the detection of a diverse set of microbes allowed the instrument cost to be paid off in three years (119). A Mayo Clinic study found that MALDI identification of yeast was faster and more cost-effective then other conventional approaches (120). Each MALDI analysis only took 5.1 minutes of hands-on time and cost only $0.50 per sample for yeast identification. In summary, studies conducted so far suggest application of MALDI for diagnosis of infections reduces the (I) time needed for analysis, (II) time patients are in the intensive care unit, (III) need for various chemicals/solvents required for conventional methods, and (IV) reduced costs in the long term (118-120). It is important to appreciate that implementation of MALDI for cell cultures on well plates (the process used in the previously-mentioned cost analysis studies) requires minimal sample preparation. By contrast, the analysis of liver biopsy tissues by MALDI requires additional steps, meaning additional cost-effectiveness studies need to be conducted. The implications of such studies for exploring the diagnostic role of MALDI on liver biopsies would be significant because there are situations in which there are no other alternative or effective techniques currently available as discussed prior. Under these circumstances, the higher cost of MALDI imaging or other low-throughput application of MALDI may be more easily justified.
One of the major factors limiting the application of MALDI to liver biopsies is that liver biopsies are routinely fixed in formalin and processed in lipophilic solvents which leads to significant loss of lipids, cross-linking of proteins and other chemical changes (78,121-123), all of which dilute the power of MALDI by artificially modifying the molecular signature of the tissue being analyzed. On the other hand, FFPWE tissues allow easier and better sectioning then other approaches for sample preservation (e.g., flash freezing), simpler long-term storage, and the maintenance of large tissue banks in pathology departments for any retrospective analysis. Thus, for clinical purposes, FFPWE preparation is attractive regarding sample storage and preservation of morphology, although loss of lipids during tissue processing makes lipidomics using MALDI less effective (122). Therefore, for the evaluation of liver diseases like fatty liver disease and certain cancers, FFPWE tissues have only limited value. On the other hand, an automated and rapid workflow has been developed and validated for the analysis of various proteins and peptides in FFPWE tissue microarrays (121). MALDI has also been applied to the evaluation of extracellular membrane proteins (including collagen and elastin) in FFPWE tissues, which are very important for evaluation of liver disease for obvious reasons (123). In addition, many metabolites (small polar molecules) have successfully been measured in FFPE tissues without any antigen retrieval or tryptic digestion, reducing the complexity of analysis (78).
Due to the chemical and physical changes induced by FFPWE, flash frozen tissues are ideal for MALDI analysis in that the molecular signature is not altered. However, frozen liver tissues during liver biopsy need to be prospectively collected, carefully stored, and it may not be always possible to keep tissue frozen due to availability of a very limited amount of tissue. In addition, frozen tissues are difficult to section, meaning that only inferior quality tissue sections may be available for overlay with MALDI images. There are additional challenges with the use of flash frozen tissues with MALDI. One major consideration is the stability of metabolites and proteins following thawing of the tissue sections. For example enzymatic activity may result in degradation of metabolites and proteins within minutes (124-126). Once matrix solvent is applied to fresh tissue sections and proteins/metabolites are extracted into the matrix, the metabolite structures are preserved to a higher extent, and the tissue sample can be stored in a vacuum sealed chamber (to reduce oxidation) at room temperature. During storage of the tissue, volatilization of the matrix can occur, leading to reduced sensitivity or other unwanted effects. As such, immediate analysis of the tissue sections after matrix application is recommended. One solution for reducing degradation of proteins/metabolites during sample preparation and freeze-thaw cycles is to deactivate enzymes by rapid exposure of the tissue sample to heat, i.e., just below boiling point. After heat treatment, the samples can be stored for a longer period at room temperature. Such treatment has been successfully applied to the reduction of degradation of metabolites and lipids (124), drugs (125) and proteins (126,127); showing drastic reduction in degradation, and in some cases no sign of degradation.
An important consideration is that MALDI analyses generate huge amounts of data. A critical issue that must be addressed prior to the clinical implementation of MALDI is the development of user-friendly and well tested bioinformatics support. Currently, the conversion of mass spectra obtained from MALDI into relevant images, categorizing tissue types and overlaying data to histological findings, identifying distinct markers across different tissue regions and disease states, and identifying the related mass spectral signatures are a bottleneck in clinical implementation of MALDI. In addition, the large amount of data generated (often terabytes), especially in MALDI imaging, requires that the software and hardware to rapidly handle large datasets should be available in laboratories/clinics applying MALDI (121).
In summary, MALDI has not only shown great potential for liver disease prognosis, diagnosis, and study of disease mechanisms, but is already being utilized in the clinical laboratory, albeit to a relatively limited extent. As the technology and informatics solutions continue to advance, new discoveries on biomarkers and applications are determined in liver biopsies, and MALDI instruments find a place in clinical laboratories, it is expected that the role of MALDI in clinical laboratories will expand. Considering the capabilities of the approach, the application of MALDI to liver biopsies for clinical management and research appears very promising and exciting.
Role of AI and ML
AI is the concept of having machines perform tasks which previously were reserved for humans. The logical next step in the use of digital images, CHiMP images and MALDI data obtained from liver biopsies is the application of ML algorithms, an application of AI, for automated generation of diagnoses and prognoses. In the context of pathology, ML corresponds to ‘teaching’ software using large and high-quality annotated datasets, from which it can ‘learn’ and formulate diagnostic opinions on future slides. ML has now been applied to various tissue types, and appears promising in the diagnosis of numerous diseases (128,129). In the area of liver pathology, the implementation of ML has been hindered by the small number of liver biopsies performed relative to other tissue specimens collected in the pathology lab. High quality computer-based algorithms require large sets of expert pathologist-acquired annotations, or labeled data (130,131). This often requires a prohibitory amounts of pathologist-screen time. However, this barrier is being reduced by the introduction of novel methods of algorithm-training, e.g., weakly supervised and transfer learning (128,130). Computer scientists have realized that they can leverage the routine workflow of pathologists in generating diagnoses to train AI models as a replacement for manual feature extraction and annotation. For example, PAIGE.ai, a leading startup company in the field of digital pathology, has implemented weakly supervised learning to achieve a high-throughput, clinical-grade algorithm to identify prostate cancer in tissue biopsies (128). Another method to leverage existing data for model construction is through transfer learning. The general concept of transfer learning is that related (if not exactly similar) datasets have been accrued at different institutions over time, and that these datasets can be consolidated to generate more robust data for ML. Recently, a fully automated ML algorithm was developed using transfer learning that accurately determined the fibrosis stage of a series of livers from mice with chemically induced fibrosis (75). These recent advances are promising for the development of new algorithms in the comparatively low volume setting that is liver pathology.
Unfortunately, some limitations of liver biopsies will either be difficult or impossible to overcome by DIA and AI. Examples include fragmentation of liver biopsies or sampling error, both of which are not uncommon in clinical practice. Fragmented specimens are difficult to stage accurately at present, and the determination of a ‘ground truth’ from which the computer could learn would be difficult to determine. To overcome this potential limitation, ML algorithms for fibrosis should also incorporate clinical datasets which include other parameters for assessment, such as evidence of portal hypertension. Sampling error is a pre-analytical error in that the tissue collected at the time of the procedure, before it reaches the pathology lab, and may not be representative of the entire tissue. Liver fibrosis is notoriously heterogeneous and can lead to this type of error (132).
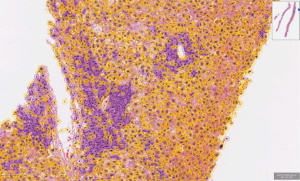
Aside from the measurement of fibrosis, the application of DIA and AI techniques to liver biopsies has been limited. ML has been used to evaluate characteristics of nuclei to differentiate benign from malignant lesions in the liver (133). From a diagnostic standpoint, there could be a role for ML in the differentiation of medical liver diseases which may appear similar upon routine histopathologic examination, such as drug-induced liver injury and autoimmune hepatitis, among others (Figure 5). For prognostic variables, there are numerous parameters which remain unexplored. For example, the amount and classification of inflammatory cells in liver biopsies of diverse etiology (e.g., neoplastic or non-neoplastic), presence and amount of bile (cholestasis), or deposited iron, are among many classifiable biomarkers that have the potential to be measured more accurately by DIA.
The reason for the overall lack of interest in applying AI to other parameters in liver pathology is complex. As previously discussed, it is at least partly due to the relatively low volume of human liver biopsies for generating an adequate amount of training datasets. There is also likely a lack of financial incentive. For-profit developers are often working to develop algorithms that pharmaceutical companies may use in clinical trials for tissue assessment. Currently, assessment of fibrosis and steatosis seem to be of most interest since there are drugs being developed to target these parameters. However, there are many other features in liver histology that are important for clinical and research purposes, but remain unexplored. When that realization has been made, an expanded role for AI in liver biopsy assessment will be developed. These considerations notwithstanding, most experts agree that AI and DIA will augment (rather than replace) the current clinical pathology workflow, at least in the near future. For example, the value of the current histologic staging systems for fibrosis is that it is pattern-based (e.g., pericentral or sinusoidal); this is not captured adequately by quantitative morphometry. Once advanced histology (e.g., CHiMP) and technology like MALDI find a role in clinical practice, it is obvious that the need for AI will increase.
Conclusions and summary
The indications for performing a liver biopsy have undergone changes in the last decade. The evolution of highly effective treatments for some of the common indications for liver biopsy in the last decade (e.g., viral hepatitis B and C) has significantly contributed to the decline in the number of liver biopsies in recent years. In addition, advanced non-invasive radiological, immunological, biochemical and genetic markers are now readily available to physicians, further diminishing or negating the need for a liver biopsy in many clinical settings. However, evaluation of liver histology remains critically important for the evaluation of a number of liver disorders and clinical settings. At the same time, the emergence of better technologies for histologic evaluation, tissue content analysis and genomics are among the many new and exciting developments in the field that hold great promise for the future and will shape the indications for a liver biopsy in the future. It is very clear that despite the advances made in the diagnosis of liver diseases, a significant proportion (5–10%) of cases remain of uncertain etiology (“cryptogenic”) and liver biopsy serves an important role in work-up of such cases. It is speculated that the vast majority of such cryptogenic cases are likely to represent inherited metabolic disorders that are either unrecognized or unusual in presentation. Application of genomics has already shown to be very helpful in work-up of such cases (134-136). The capabilities of these evolving techniques (ChiMP and MALDI) as discussed above go far beyond routine lipidomics and proteomics. We anticipate that their application in clinical practice is also likely to contribute significantly in the understanding and diagnosis of liver diseases of uncertain etiology.
Acknowledgments
We thank our colleagues, especially Dr. David C. Thompson, for valuable discussions and critical reading of the manuscript.
Funding: This work was partly supported by NIH grants R44 CA189522-01, AA017754 and AA022057.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Luca Fabris and Mario Strazzabosco) for the series “Recent Advances in Rare Liver Diseases” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh.2020.04.01). The series “Recent Advances in Rare Liver Diseases” was commissioned by the editorial office without any funding or sponsorship. Dr. VV reports grants from NIAAA/NIH (AA017754 and AA022057), during the conduct of the study; Dr. RT reports grant support from National Institutes of Health (NCI, R44 CA189522-01), during the conduct of the study; In addition, Dr. RT has an ownership interest in Applikate Technologies, a startup that is commercializing multiphoton histology, and related intellectual property. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Menghini G. One-second needle biopsy of the liver. Gastroenterology 1958;35:190-9. [Crossref] [PubMed]
- Sheela H, Seela S, Caldwell C, et al. Liver biopsy: evolving role in the new millennium. Journal of Clinical Gastroenterology 2005;39:603-10. [Crossref] [PubMed]
- Ravindran S, Hancox SH, Howlett DC. Liver biopsy: past, present and future. Br J Hosp Med (Lond) 2016;77:90-5. [Crossref] [PubMed]
- Beckmann MG, Bahr MJ, Hadem J, et al. Clinical relevance of transjugular liver biopsy in comparison with percutaneous and laparoscopic liver biopsy. Gastroenterol Res Pract 2009;2009:947014. [Crossref] [PubMed]
- Rodziewicz M, Moreno-Otero R. Role of percutaneous liver biopsy. Hepat Mon 2012;12:294-5. [Crossref] [PubMed]
- Dohan A, Guerrache Y, Boudiaf M, et al. Transjugular liver biopsy: indications, technique and results. Diagn Interv Imaging 2014;95:11-5. [Crossref] [PubMed]
- Falcone RE, Wanamaker SR, Barnes F, et al. Laparoscopic vs. open wedge biopsy of the liver. J Laparoendosc Surg 1993;3:325-9. [Crossref] [PubMed]
- Padoin AV, Mottin CC, Moretto M, et al. A comparison of wedge and needle hepatic biopsy in open bariatric surgery. Obes Surg 2006;16:178-82. [Crossref] [PubMed]
- Arab JP, Barrera F, Arrese M. The Evolving Role of Liver Biopsy in Non-alcoholic Fatty Liver Disease. Ann Hepatol 2018;17:899-902. [Crossref] [PubMed]
- Kheir AE, Ahmed WM, Gaber I, et al. Idiopathic neonatal hepatitis or extrahepatic biliary atresia? The role of liver biopsy. Sudan J Paediatr 2016;16:58-62. [PubMed]
- Cadranel JF, Nousbaum JB, Gouillou M, et al. Major changes in the number and indications of liver biopsy for chronic liver diseases over one decade in France. Eur J Gastroenterol Hepatol 2016;28:e26-32. [Crossref] [PubMed]
- Amarapurkar D, Amarapurkar A. Indications of Liver Biopsy in the Era of Noninvasive Assessment of Liver Fibrosis. J Clin Exp Hepatol 2015;5:314-9. [Crossref] [PubMed]
- Tannapfel A, Dienes HP, Lohse AW. The indications for liver biopsy. Dtsch Arztebl Int 2012;109:477-83. [PubMed]
- Lipp MJ, D'Souza LS, Clain DJ, et al. Trends in the indication and method of liver biopsy for hepatitis B and C. Dig Dis Sci 2010;55:2971-6. [Crossref] [PubMed]
- Saadeh S, Cammell G, Carey WD, et al. The role of liver biopsy in chronic hepatitis C. Hepatology 2001;33:196-200. [Crossref] [PubMed]
- Mani H, Kleiner DE. Liver biopsy findings in chronic hepatitis B. Hepatology 2009;49:S61-71. [Crossref] [PubMed]
- Friedman SL, Bansal MB. Reversal of hepatic fibrosis -- fact or fantasy? Hepatology 2006;43:S82-8. [Crossref] [PubMed]
- Pérez-Tamayo R. Cirrhosis of the liver: a reversible disease? Pathol Annu 1979;14:183-213. [PubMed]
- Wanless IR, Nakashima E, Sherman M. Regression of human cirrhosis. Morphologic features and the genesis of incomplete septal cirrhosis. Arch Pathol Lab Med 2000;124:1599-607. [PubMed]
- Serpaggi J, Carnot F, Nalpas B, et al. Direct and indirect evidence for the reversibility of cirrhosis. Hum Pathol 2006;37:1519-26. [Crossref] [PubMed]
- Knodell RG, Ishak KG, Black WC, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1981;1:431-5. [Crossref] [PubMed]
- Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 1996;24:289-93. [Crossref] [PubMed]
- Bedossa P, Garcia-Tsao G, Jain D. Cirrhosis Regression and Subclassification. Surg Pathol Clin 2013;6:295-309. [Crossref] [PubMed]
- Kim SU, Oh HJ, Wanless IR, et al. The Laennec staging system for histological sub-classification of cirrhosis is useful for stratification of prognosis in patients with liver cirrhosis. J Hepatol 2012;57:556-63. [Crossref] [PubMed]
- Tsochatzis E, Bruno S, Isgro G, et al. Collagen proportionate area is superior to other histological methods for sub-classifying cirrhosis and determining prognosis. J Hepatol 2014;60:948-54. [Crossref] [PubMed]
- Hytiroglou P, Theise ND. Regression of human cirrhosis: an update, 18 years after the pioneering article by Wanless et al. Virchows Arch 2018;473:15-22. [Crossref] [PubMed]
- D'Ambrosio R, Aghemo A, Rumi MG, et al. Histological findings among HCV cirrhotic patients who achieved a SVR: a morphometric and immunohistochemical study. Hepatology 2011;54:1229A-A.
- Sun Y, Zhou J, Wang L, et al. New classification of liver biopsy assessment for fibrosis in chronic hepatitis B patients before and after treatment. Hepatology 2017;65:1438-50. [Crossref] [PubMed]
- Theise ND, Jia J, Sun Y, et al. Progression and regression of fibrosis in viral hepatitis in the treatment era: the Beijing classification. Mod Pathol 2018;31:1191-200. [Crossref] [PubMed]
- Rashidi HH, Tran NK, Betts EV, et al. Artificial Intelligence and Machine Learning in Pathology: The Present Landscape of Supervised Methods. Acad Pathol 2019;6:2374289519873088. [Crossref] [PubMed]
- Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017;318:2199-210. [Crossref] [PubMed]
- Jimenez W, Pares A, Caballeria J, et al. Measurement of fibrosis in needle liver biopsies: evaluation of a colorimetric method. Hepatology 1985;5:815-8. [Crossref] [PubMed]
- Calvaruso V, Burroughs AK, Standish R, et al. Computer-assisted image analysis of liver collagen: relationship to Ishak scoring and hepatic venous pressure gradient. Hepatology 2009;49:1236-44. [Crossref] [PubMed]
- Friedenberg MA, Miller L, Chung CY, et al. Simplified method of hepatic fibrosis quantification: design of a new morphometric analysis application. Liver Int 2005;25:1156-61. [Crossref] [PubMed]
- Pilette C, Rousselet MC, Bedossa P, et al. Histopathological evaluation of liver fibrosis: quantitative image analysis vs semi-quantitative scores. Comparison with serum markers. J Hepatol 1998;28:439-46. [Crossref] [PubMed]
- Masseroli M, Caballero T, O'Valle F, et al. Automatic quantification of liver fibrosis: design and validation of a new image analysis method: comparison with semi-quantitative indexes of fibrosis. J Hepatol 2000;32:453-64. [Crossref] [PubMed]
- Zhou Y, Ru GQ, Yan R, et al. An Inexpensive Digital Image Analysis Technique for Liver Fibrosis Quantification in Chronic Hepatitis B Patients. Ann Hepatol 2017;16:881-7. [Crossref] [PubMed]
- Caballero T, Perez-Milena A, Masseroli M, et al. Liver fibrosis assessment with semiquantitative indexes and image analysis quantification in sustained-responder and non-responder interferon-treated patients with chronic hepatitis C. J Hepatol 2001;34:740-7. [Crossref] [PubMed]
- Maduli E, Andorno S, Rigamonti C, et al. Evaluation of liver fibrosis in chronic hepatitis C with a computer-assisted morphometric method. Ann Ital Med Int 2002;17:242-7. [PubMed]
- Boursier J, Chaigneau J, Roullier V, et al. Steatosis degree, measured by morphometry, is linked to other liver lesions and metabolic syndrome components in patients with NAFLD. Eur J Gastroenterol Hepatol 2011;23:974-81. [Crossref] [PubMed]
- Lin J, Lu F, Zheng W, et al. Assessment of liver steatosis and fibrosis in rats using integrated coherent anti-Stokes Raman scattering and multiphoton imaging technique. J Biomed Opt 2011;16:116024. [Crossref] [PubMed]
- Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005;41:1313-21. [Crossref] [PubMed]
- Marsman WA, Wiesner RH, Rodriguez L, et al. Use of fatty donor liver is associated with diminished early patient and graft survival. Transplantation 1996;62:1246-51. [Crossref] [PubMed]
- Marsman H, Matsushita T, Dierkhising R, et al. Assessment of donor liver steatosis: pathologist or automated software? Hum Pathol 2004;35:430-5. [Crossref] [PubMed]
- Hall AR, Dhillon AP, Green AC, et al. Hepatic steatosis estimated microscopically versus digital image analysis. Liver Int 2013;33:926-35. [Crossref] [PubMed]
- Franzén LE, Ekstedt M, Kechagias S, et al. Semiquantitative evaluation overestimates the degree of steatosis in liver biopsies: a comparison to stereological point counting. Mod Pathol 2005;18:912-6. [Crossref] [PubMed]
- Mancini M, Summers P, Faita F, et al. Digital liver biopsy: Bio-imaging of fatty liver for translational and clinical research. World J Hepatol 2018;10:231-45. [Crossref] [PubMed]
- Liao J, Jiang Y, Bian Z, et al. Rapid focus map surveying for whole slide imaging with continuous sample motion. Opt Lett 2017;42:3379-82. [Crossref] [PubMed]
- Gilbertson JR, Ho J, Anthony L, et al. Primary histologic diagnosis using automated whole slide imaging: a validation study. BMC Clin Pathol 2006;6:4. [Crossref] [PubMed]
- Massone C, Soyer HP, Lozzi GP, et al. Feasibility and diagnostic agreement in teledermatopathology using a virtual slide system. Hum Pathol 2007;38:546-54. [Crossref] [PubMed]
- Liang Y, Shilagard T, Xiao SY, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology 2009;137:1448-58. [Crossref] [PubMed]
- Protiva P, Gong J, Sreekumar B, et al. Pigment Epithelium-Derived Factor (PEDF) Inhibits Wnt/beta-catenin Signaling in the Liver. Cell Mol Gastroenterol Hepatol 2015;1:535-49.e14. [Crossref] [PubMed]
- Lee HS, Liu Y, Chen HC, et al. Optical biopsy of liver fibrosis by use of multiphoton microscopy. Opt Lett 2004;29:2614-6. [Crossref] [PubMed]
- Zhu Y, Gao W, Zhou Y, et al. Rapid and high-resolution imaging of human liver specimens by full-field optical coherence tomography. J Biomed Opt 2015;20:116010. [Crossref] [PubMed]
- Satoh S, Otsuka Y, Ozeki Y, et al. Label‐free visualization of acetaminophen‐induced liver injury by high‐speed stimulated R aman scattering spectral microscopy and multivariate image analysis. Pathol Int 2014;64:518-26. [Crossref] [PubMed]
- Pampaloni F, Chang BJ, Stelzer EHK. Light sheet-based fluorescence microscopy (LSFM) for the quantitative imaging of cells and tissues. Cell Tissue Res 2015;360:129-41. [Crossref] [PubMed]
- Fereidouni F, Mitra AD, Demos S, et al. Microscopy with UV Surface Excitation (MUSE) for slide-free histology and pathology imaging. BiOS SPIE, 2015.
- Campo-Ruiz V, Ochoa ER, Lauwers GY, et al. Evaluation of hepatic histology by near-infrared confocal microscopy: a pilot study. Hum Pathol 2002;33:975-82. [Crossref] [PubMed]
- Wang H, Liang X, Gravot G, et al. Visualizing liver anatomy, physiology and pharmacology using multiphoton microscopy. J Biophotonics 2017;10:46-60. [Crossref] [PubMed]
- Torres R, Vesuna S, Levene MJ. High-Resolution, 2- and 3-Dimensional Imaging of Uncut, Unembedded Tissue Biopsy Samples. Arch Pathol Lab Med 2014;138:395-402. [Crossref] [PubMed]
- Glaser AK, Reder NP, Chen Y, et al. Light-sheet microscopy for slide-free non-destructive pathology of large clinical specimens. Nat Biomed Eng 2017;1:0084.
- Yu T, Qi Y, Gong H, et al. Optical clearing for multiscale biological tissues. J Biophotonics 2018;11:e201700187. [Crossref] [PubMed]
- Campo-Ruiz V, Lauwers GY, Anderson RR, et al. In vivo and ex vivo virtual biopsy of the liver with near-infrared, reflectance confocal microscopy. Mod Pathol 2005;18:290. [Crossref] [PubMed]
- Thorling CA, Crawford D, Burczynski FJ, et al. Multiphoton microscopy in defining liver function. J Biomed Opt 2014;19:90901. [Crossref] [PubMed]
- Eliceiri KW, Fan CH, Lyons GE, et al. Analysis of histology specimens using lifetime multiphoton microscopy. J Biomed Opt 2003;8:376-80. [Crossref] [PubMed]
- Yan J, Jiang J, Jia F, et al. Preclinical study of using multiphoton microscopy to diagnose liver cancer and differentiate benign and malignant liver lesions. J Biomed Opt 2012;17:026004. [Crossref] [PubMed]
- Wang H, Liang X, Mohammed YH, et al. Real-time histology in liver disease using multiphoton microscopy with fluorescence lifetime imaging. Biomed Opt Express 2015;6:780-92. [Crossref] [PubMed]
- Sun W, Chang S, Tai DC, et al. Nonlinear optical microscopy: use of second harmonic generation and two-photon microscopy for automated quantitative liver fibrosis studies. J Biomed Opt 2008;13:064010. [Crossref] [PubMed]
- Sun TL, Liu Y, Sung M-C, et al. Ex vivo imaging and quantification of liver fibrosis using second-harmonic generation microscopy. J Biomed Opt 2010;15:036002. [Crossref] [PubMed]
- Gailhouste L, Le Grand Y, Odin C, et al. Fibrillar collagen scoring by second harmonic microscopy: a new tool in the assessment of liver fibrosis. J Hepatol 2010;52:398-406. [Crossref] [PubMed]
- Halász T, Horváth G, Kiss A, et al. Evaluation of Histological and non-Invasive Methods for the Detection of Liver Fibrosis: The Values of Histological and Digital Morphometric Analysis, Liver Stiffness Measurement and APRI Score. Pathol Oncol Res 2016;22:1-6. [Crossref] [PubMed]
- Olson E, Levene MJ, Torres R. Multiphoton microscopy with clearing for three dimensional histology of kidney biopsies. Biomed Opt Express 2016;7:3089-96. [Crossref] [PubMed]
- Arjmand A, Angelis CT, Tzallas AT, et al. editors. Deep Learning in Liver Biopsies using Convolutional Neural Networks. 2019 42nd International Conference on Telecommunications and Signal Processing (TSP), 2019.
- Li CT, Chung PC, Tsai HW, et al. editors. Inflammatory Cells Detection in H&E Staining Histology Images Using Deep Convolutional Neural Network with Distance Transformation. International Computer Symposium; Springer, 2018.
- Yu Y, Wang J, Ng CW, et al. Deep learning enables automated scoring of liver fibrosis stages. Sci Rep 2018;8:16016. [Crossref] [PubMed]
- Lin H, Wei C, Wang G, et al. Automated classification of hepatocellular carcinoma differentiation using multiphoton microscopy and deep learning. J Biophotonics 2019;12:e201800435. [Crossref] [PubMed]
- Charkoftaki G, Rattray NJW, Andren PE, et al. Yale School of Public Health Symposium on tissue imaging mass spectrometry: illuminating phenotypic heterogeneity and drug disposition at the molecular level. Hum Genomics 2018;12:10. [Crossref] [PubMed]
- Ly A, Buck A, Balluff B, et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat Protoc 2016;11:1428-43. [Crossref] [PubMed]
- Ucal Y, Durer ZA, Atak H, et al. Clinical applications of MALDI imaging technologies in cancer and neurodegenerative diseases. Biochim Biophys Acta Proteins Proteom 2017;1865:795-816. [Crossref] [PubMed]
- Balluff B, Rauser S, Meding S, et al. MALDI imaging identifies prognostic seven-protein signature of novel tissue markers in intestinal-type gastric cancer. Am J Pathol 2011;179:2720-9. [Crossref] [PubMed]
- Casadonte R, Kriegsmann M, Zweynert F, et al. Imaging mass spectrometry to discriminate breast from pancreatic cancer metastasis in formalin-fixed paraffin-embedded tissues. Proteomics 2014;14:956-64. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Thomas A, Patterson NH, Marcinkiewicz MM, et al. Histology-driven data mining of lipid signatures from multiple imaging mass spectrometry analyses: application to human colorectal cancer liver metastasis biopsies. Anal Chem 2013;85:2860-6. [Crossref] [PubMed]
- Park ES, Lee JH, Hong JH, et al. Phosphatidylcholine alteration identified using MALDI imaging MS in HBV-infected mouse livers and virus-mediated regeneration defects. PLoS One 2014;9:e103955. [Crossref] [PubMed]
- Cobo F. Application of maldi-tof mass spectrometry in clinical virology: a review. Open Virol J 2013;7:84-90. [Crossref] [PubMed]
- Ščupáková K, Soons Z, Ertaylan G, et al. Spatial Systems Lipidomics Reveals Nonalcoholic Fatty Liver Disease Heterogeneity. Anal Chem 2018;90:5130-8. [Crossref] [PubMed]
- Lièvre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res 2006;66:3992-5. [Crossref] [PubMed]
- Schieving JH, de Vries M, van Vugt JM, et al. Alpha-fetoprotein, a fascinating protein and biomarker in neurology. Eur J Paediatr Neurol 2014;18:243-8. [Crossref] [PubMed]
- Han EC, Lee YS, Liao WS, et al. Direct tissue analysis by MALDI-TOF mass spectrometry in human hepatocellular carcinoma. Clin Chim Acta 2011;412:230-9. [Crossref] [PubMed]
- Cuccurullo V, Di Stasio GD, Mazzarella G, et al. Microvascular Invasion in HCC: The Molecular Imaging Perspective. Contrast Media Mol Imaging 2018;2018:9487938. [Crossref] [PubMed]
- Poté N, Alexandrov T, Le Faouder J, et al. Imaging mass spectrometry reveals modified forms of histone H4 as new biomarkers of microvascular invasion in hepatocellular carcinomas. Hepatology 2013;58:983-94. [Crossref] [PubMed]
- Le Faouder J, Laouirem S, Chapelle M, et al. Imaging mass spectrometry provides fingerprints for distinguishing hepatocellular carcinoma from cirrhosis. J Proteome Res 2011;10:3755-65. [Crossref] [PubMed]
- Le Faouder J, Laouirem S, Alexandrov T, et al. Tumoral heterogeneity of hepatic cholangiocarcinomas revealed by MALDI imaging mass spectrometry. Proteomics 2014;14:965-72. [Crossref] [PubMed]
- Marquardt C, Tolstik T, Bielecki C, et al. MALDI imaging-based classification of hepatocellular carcinoma and non-malignant lesions in fibrotic liver tissue. Z Gastroenterol 2015;53:33-9. [Crossref] [PubMed]
- Meding S, Nitsche U, Balluff B, et al. Tumor classification of six common cancer types based on proteomic profiling by MALDI imaging. J Proteome Res 2012;11:1996-2003. [Crossref] [PubMed]
- Morita Y, Sakaguchi T, Ikegami K, et al. Lysophosphatidylcholine acyltransferase 1 altered phospholipid composition and regulated hepatoma progression. J Hepatol 2013;59:292-9. [Crossref] [PubMed]
- Lim YS, Kim WR. The global impact of hepatic fibrosis and end-stage liver disease. Clin Liver Dis 2008;12:733-46. vii. [Crossref] [PubMed]
- Singh S, Osna NA, Kharbanda KK. Treatment options for alcoholic and non-alcoholic fatty liver disease: A review. World J Gastroenterol 2017;23:6549-70. [Crossref] [PubMed]
- Toshikuni N, Tsutsumi M, Arisawa T. Clinical differences between alcoholic liver disease and nonalcoholic fatty liver disease. World J Gastroenterol 2014;20:8393-406. [Crossref] [PubMed]
- Hamer OW, Aguirre DA, Casola G, et al. Fatty liver: imaging patterns and pitfalls. Radiographics 2006;26:1637-53. [Crossref] [PubMed]
- Sakhuja P. Pathology of alcoholic liver disease, can it be differentiated from nonalcoholic steatohepatitis? World J Gastroenterol 2014;20:16474-9. [Crossref] [PubMed]
- Hall Z, Bond NJ, Ashmore T, et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology 2017;65:1165-80. [Crossref] [PubMed]
- Alamri H, Patterson NH, Yang E, et al. Mapping the triglyceride distribution in NAFLD human liver by MALDI imaging mass spectrometry reveals molecular differences in micro and macro steatosis. Anal Bioanal Chem 2019;411:885-94. [Crossref] [PubMed]
- Nishikawa K, Hashimoto M, Itoh Y, et al. Detection of changes in the structure and distribution map of triacylglycerol in fatty liver model by MALDI-SpiralTOF. FEBS Open Bio 2014;4:179-84. [Crossref] [PubMed]
- Rasineni K, Casey CA. Molecular mechanism of alcoholic fatty liver. Indian J Pharmacol 2012;44:299-303. [Crossref] [PubMed]
- Régnier M, Polizzi A, Guillou H, et al. Sphingolipid metabolism in non-alcoholic fatty liver diseases. Biochimie 2019;159:9-22. [Crossref] [PubMed]
- Feldstein AE, Lopez R, Tamimi TA, et al. Mass spectrometric profiling of oxidized lipid products in human nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J Lipid Res 2010;51:3046-54. [Crossref] [PubMed]
- Pritchard MT, McMullen MR, Stavitsky AB, et al. Differential contributions of C3, C5, and decay-accelerating factor to ethanol-induced fatty liver in mice. Gastroenterology 2007;132:1117-26. [Crossref] [PubMed]
- Loftus N, Barnes A, Ashton S, et al. Metabonomic investigation of liver profiles of nonpolar metabolites obtained from alcohol-dosed rats and mice using high mass accuracy MSn analysis. J Proteome Res 2011;10:705-13. [Crossref] [PubMed]
- Schröck A, Henzi A, Butikofer P, et al. Determination of the formation rate of phosphatidylethanol by phospholipase D (PLD) in blood and test of two selective PLD inhibitors. Alcohol 2018;73:1-7. [Crossref] [PubMed]
- Gorden DL, Myers DS, Ivanova PT, et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res 2015;56:722-36. [Crossref] [PubMed]
- Talwani R, Gilliam BL, Howell C. Infectious diseases and the liver. Clin Liver Dis 2011;15:111-30. [Crossref] [PubMed]
- Garner O, Mochon A, Branda J, et al. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK(R) MS system. Clin Microbiol Infect 2014;20:335-9. [Crossref] [PubMed]
- Rychert J, Burnham CA, Bythrow M, et al. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J Clin Microbiol 2013;51:2225-31. [Crossref] [PubMed]
- Schulthess B, Bloemberg GV, Zbinden R, et al. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J Clin Microbiol 2014;52:1089-97. [Crossref] [PubMed]
- Marko DC, Saffert RT, Cunningham SA, et al. Evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of nonfermenting gram-negative bacilli isolated from cultures from cystic fibrosis patients. J Clin Microbiol 2012;50:2034-9. [Crossref] [PubMed]
- Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin Chem 2015;61:100-11. [Crossref] [PubMed]
- Patel TS, Kaakeh R, Nagel JL, et al. Cost Analysis of Implementing Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Plus Real-Time Antimicrobial Stewardship Intervention for Bloodstream Infections. J Clin Microbiol 2016;55:60-7. [Crossref] [PubMed]
- Tran A, Alby K, Kerr A, et al. Cost Savings Realized by Implementation of Routine Microbiological Identification by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J Clin Microbiol 2015;53:2473-9. [Crossref] [PubMed]
- Dhiman N, Hall L, Wohlfiel SL, et al. Performance and cost analysis of matrix-assisted laser desorption ionization-time of flight mass spectrometry for routine identification of yeast. J Clin Microbiol 2011;49:1614-6. [Crossref] [PubMed]
- Groseclose MR, Massion PP, Chaurand P, et al. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 2008;8:3715-24. [Crossref] [PubMed]
- Wisztorski M, Franck J, Salzet M, et al. MALDI direct analysis and imaging of frozen versus FFPE tissues: what strategy for which sample? Methods Mol Biol 2010;656:303-22. [Crossref] [PubMed]
- Angel PM, Comte-Walters S, Ball LE, et al. Mapping Extracellular Matrix Proteins in Formalin-Fixed, Paraffin-Embedded Tissues by MALDI Imaging Mass Spectrometry. J Proteome Res 2018;17:635-46. [Crossref] [PubMed]
- Koelmel JP, Jones CM, Ulmer CZ, et al. Examining heat treatment for stabilization of the lipidome. Bioanalysis 2018;10:291-305. [Crossref] [PubMed]
- Blessborn D, Skold K, Zeeberg D, et al. Heat stabilization of blood spot samples for determination of metabolically unstable drug compounds. Bioanalysis 2013;5:31-9. [Crossref] [PubMed]
- Goodwin RJ, Lang AM, Allingham H, et al. Stopping the clock on proteomic degradation by heat treatment at the point of tissue excision. Proteomics 2010;10:1751-61. [Crossref] [PubMed]
- Kultima K, Skold K, Boren M. Biomarkers of disease and post-mortem changes - Heat stabilization, a necessary tool for measurement of protein regulation. J Proteomics 2011;75:145-59. [Crossref] [PubMed]
- Campanella G, Hanna MG, Geneslaw L, et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med 2019;25:1301-9. [Crossref] [PubMed]
- Zhang XL, Wang D. A Deep Ensemble Learning Method for Monaural Speech Separation. IEEE/ACM Trans Audio Speech Lang Process 2016;24:967-77. [Crossref] [PubMed]
- Xie J, Liu R, Luttrell JT, et al. Deep Learning Based Analysis of Histopathological Images of Breast Cancer. Front Genet 2019;10:80. [Crossref] [PubMed]
- Fuchs TJ, Buhmann JM. Computational pathology: challenges and promises for tissue analysis. Comput Med Imaging Graph 2011;35:515-30. [Crossref] [PubMed]
- Bedossa P, Dargere D, Paradis V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology 2003;38:1449-57. [Crossref] [PubMed]
- Xia R, Boroujeni AM, Shea S, et al. Diagnosis of Liver Neoplasms by Computational and Statistical Image Analysis. Gastroenterology Res 2019;12:288-98. [Crossref] [PubMed]
- Gao E, Cheema H, Waheed N, et al. OSTalpha deficiency: A disorder with cholestasis, liver fibrosis and congenital diarrhea. Hepatology 2020;71:1879-82. [Crossref] [PubMed]
- Hakim A, Zhang X, DeLisle A, et al. Clinical utility of genomic analysis in adults with idiopathic liver disease. J Hepatol 2019;70:1214-21. [Crossref] [PubMed]
- Vilarinho S, Mistry PK. Exome Sequencing in Clinical Hepatology. Hepatology 2019;70:2185-92. [Crossref] [PubMed]
Cite this article as: Jain D, Torres R, Celli R, Koelmel J, Charkoftaki G, Vasiliou V. Evolution of the liver biopsy and its future. Transl Gastroenterol Hepatol 2021;6:20.


