
Genetic and epigenetic disease modifiers: non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD)
Introduction
Non-alcoholic fatty liver disease (NAFLD) has become the most common liver disease, the world over and encompasses a spectrum ranging from simple steatosis (non-alcoholic fatty liver or NAFL), non-alcoholic steatohepatitis (NASH), NASH related cirrhosis and hepatocellular carcinoma (HCC) (1,2). In some parts of the world, equally common problem is the alcoholic liver disease (ALD) with a similar spectrum of alcoholic steatosis, alcoholic steatohepatitis, cirrhosis and HCC (3). There are significant inter-individual differences in the severity and progression of liver disease in patients with NAFLD sharing similar metabolic and other environmental risk factors (4). Similarly severity of liver disease differs among patients consuming similar type and quantity of alcohol over a similar duration (5). In addition to inter-individual differences, inter-ethnic differences in NAFLD and ALD and familial clustering points towards the role of genetic and epigenetic factors in the pathogenesis of these two diseases (6-11). In this review article, we discuss the role of genetic and epigenetic modifiers in NAFLD and ALD.
Genetic basis in the pathogenesis of NAFLD
NAFLD is a complex disease with multiple factors involved in the pathogenesis that include both environmental and genetic modifiers. Also, NASH and fibrosis progression varies with some patients being ‘rapid progressors’ (12,13). Risk factors for fibrosis progression in patients with NAFLD include metabolic syndrome, presence of type 2 diabetes mellitus, high body mass index (BMI)/obesity, age, steatosis grade, high insulin resistance, baseline biopsy showing inflammation and fibrosis (1,2,13). In a meta-analysis of studies including paired biopsies, Singh et al. showed that rate of fibrosis progression was twice as common in patients with NASH in comparison to patients with NAFL (12).
In addition to environmental and metabolic risk factors, familial clustering, studies involving the monozygotic twins and inter-ethnic differences point towards the role of genetics in the pathogenesis of NAFLD.
Familial clustering in NAFLD
Several studies have shown the clustering of cases of NAFLD among the families (8,14,15). Brouwers et al. compared 157 familial combined hyperlipidemia family members with 20 spouses. Fatty liver was more prevalent in probands (40%) and relatives (35%) compared to spouses (15%) (8). In a study, siblings and parents of overweight/obese children with and without NAFLD were compared with the help of magnetic resonance proton density fat fraction (MR-PDFF) (15). NAFLD was present in 17% of siblings and 37% of parents in non-NAFLD overweight/obese children group in comparison to presence of NAFLD in 59% of siblings and 78% of parents of overweight/obese children with NAFLD. The correlation of liver fat fraction to BMI was stronger in families of children with NAFLD than without NAFLD. The authors found heritability of NAFLD as 1.000 and that of liver fat fraction as 0.386 (15).
Twin studies
Prevalence studies in twins suggest a stronger genetic basis of NAFLD in comparison to that evident from familial clustering studies. Several twin studies have shown higher prevalence of NAFLD in monozygotic twins as compared to dizygotic twins. Makkonen et al. studied 313 twin pairs and observed significantly higher intra-pair correlations in monozygotic than the dizygotic twins for both alanine transaminases (ALT) (0.65 and 0.04 respectively) and fasting serum insulin (0.58 and 0.34 respectively). Heritability of ALT was 55% and that of fasting serum insulin was 61%. ALT and fasting serum insulin correlated with liver fat content in 66 subjects (measured by magnetic resonance spectroscopy) (16). In a study using ultrasonography for NAFLD and carotid intima media thickness in 208 (58 men and 150 women) Hungarian twins (63 monozygotic and 41 dizygotic pairs, aged 43.7±16.7 years), heritability was linked to cardiovascular risk, but not to NAFLD (17). However, it should be noted that ultrasound with its limitations may have categorized some patients with mild steatosis as normal on ultrasound. Loomba et al. studied 60 pairs of twins (42 monozygotic and 18 dizygotic) aged 45.7±22.1 years. Steatosis and fibrosis were measured by MR-PDFF and MR elastography (MRE) in the study subjects. Twenty-six (21.7%) had NAFLD, which correlated between monozygotic twins (r2=0.70; P<0.0001) but not in dizygotic twins. The fibrosis also correlated between monozygotic twins (r2=0.48; P<0.002) but not in dizygotic twins (r2=0.12; P=0.7). The heritability of hepatic steatosis was 0.52 and of hepatic fibrosis was 0.5 (18). In a prospective study, Cui et al. studied 65 twin pairs (45 monozygotic, 20 dizygotic twin pairs, aged 47.1±21.9 years) and found that 20% (n=26) had hepatic steatosis and 8.2% (n=10) had hepatic fibrosis. Steatosis and fibrosis had a significant shared gene effect of 0.756 (95% CI, 0.716–1) (19).
Ethnic differences
Major evidence for having the genetic basis in NAFLD comes from studies showing the ethnic differences in the prevalence and severity of NAFLD. In a study including cryptogenic cirrhosis or NASH related cirrhosis, it was seen that prevalence of cirrhosis among Hispanic and African American patients was 3.1 times higher and 3.9 times lower than European American patients despite similar prevalence of type 2 diabetes mellitus among these groups (20). Wagenknecht et al. compared 795 Hispanic-American and 347 African-American adults, aged 49 [22–84] years. NAFLD was present in 24% of Hispanics versus 10% in African-American diagnosed on the basis of CT scan. NAFLD was independently associated with insulin sensitivity and visceral adipose tissue area in both ethnic groups, proportion of explained variance being higher in Hispanics (21). Browning et al. studied liver steatosis by proton magnetic resonance spectroscopy in 2,287 patients of multi-ethnic population comprising of 32.1% Whites, 48.3% African Americans and 17.5% Hispanics. NAFLD was present in 45% of Hispanics; 33% of Whites and 24% of African Americans. Higher prevalence of NAFLD in Hispanics in comparison to African Americans in spite of the presence of metabolic risk factors in later group suggested the genetic basis for the difference (6). A study analyzing 1,026 adults with biopsy proven NAFLD from the Nonalcoholic Steatohepatitis Clinical Research Network observed that HOMA-IR was not a significant risk factor for NASH among Latinos, but was significant among non-Latino Whites (22). In a systemic review of 34 studies (368,569 patients), NAFLD prevalence was highest in the Hispanics (relative risk 1.09) and lower in African Americans (relative risk 0.72) in comparison to Whites, with no difference in hepatic fibrosis (7). While most of data regarding ethnic differences is available in non-Asians, few studies have addressed this issue in the Asian population. Mohanty et al. showed that Asians had higher grades of histological ballooning in comparison to NAFLD patients of other ethnicities (OR 2.71, P=0.04). Hispanics on the other hand showed a higher prevalence of Mallory hyaline in comparison to patients of other ethnicities (OR 2.41, P=0.03) and African Americans had lower degree of hepatic steatosis (23). In another study from John Hopkins Hospital, Baltimore, Maryland, authors observed severe hepatic steatosis and a trend towards severe inflammation in Asian-Americans (24). Petersen et al. compared results of oral glucose tolerance test in Caucasians (n=292), Asian-Indians (n=59), Eastern Asians (n=49), African Americans (n=48) and Hispanics (n=34). The prevalence of insulin resistance was 2–3-fold higher in the Asian-Indians as compared to other ethnicities. There was approximately 2-fold increase in hepatic triglyceride content in Asians as compared to Caucasian men (25). The difference in the prevalence and severity among patients with different ethnicities is best explained by the difference in various gene variants.
Genetic modifiers in NAFLD
Two types of genetic studies are available in patients with NAFLD. Candidate gene studies are hypothesis-testing studies, which are done for a gene with known functions. Candidate gene studies look for difference regarding a polymorphism in cases and controls. A small sample size is needed for candidate gene studies but these studies are not able to find new genetic associations. On the other hand, genome wide association studies (GWAS) are hypothesis-generating studies. GWAS look at thousands to millions of short nucleotide polymorphisms (SNPs). GWAS are done in a large sample size, and are useful to find new genetic associations of a disease. GWAS studies in NAFLD are shown in Table 1 and candidate gene studies are shown in Table 2.
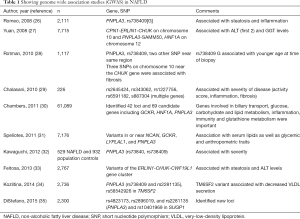
Full table
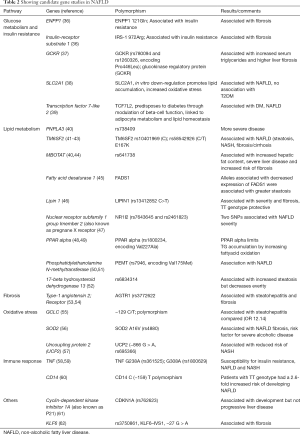
Full table
Genetic association of NAFLD with PNPLA3, TM6SF2 and other gene variants
The strongest evidence for the genetic link in NAFLD has been shown to be related to Patatin-like phospholipase domain-containing protein 3 (PNPLA3) also known as adiponutrin (ADPN), acylglycerol O-acyltransferase or calcium-independent phospholipase A2-epsilon (iPLA2-epsilon), an enzyme that in humans is encoded by the PNPLA3 gene located on chromosome 22. The PNPLA3 variant (rs738409 C > G, p.I148M) is a cytosine to guanine nucleotide transversion mutation at codon 148 that causes isoleucine to methionine amino acid change (63) and has been shown to be strongly associated with increased hepatic steatosis. PNPLA3 or Adiponutrin is a triacylglycerol lipase that possesses both lipolytic and lipogenic activities in vitro.
The I148M substitution has been shown to cause both gain and loss of function; loss of function by decreasing lipolytic activity, thus increasing triglyceride accumulation (64). Pirazzi et al. studied very-low-density lipoprotein (VLDL) kinetics in 55 overweight/obese men. The authors showed that PNPLA3 I148M variant affected secretion of apoB-containing lipoproteins, suggestive of a loss-of-function mutation. The authors suggested that PNPLA3 148M promoted intracellular lipid accumulation by reducing the lipidation of VLDL (65). There is also evidence of increased lipid synthesis by I148M variant. In a study from Austria, it was shown that PNPLA3 also acts as acyl-CoA-dependent lysophosphatidic acid acyltransferase (LPAAT); I148M variant showed elevated LPAAT activity in comparison to the wild-type, thus promoting hepatic lipid synthesis by gain of function (66). PNPLA3 also affects hepatic stellate cells (HSCs), which play an important role in the development of liver fibrosis. HSCs are the main reservoir of the retinoids. Upon activation, HSCs lose retinol content and differentiate into activated myofibroblasts, which produce collagen (fibrosis). PNPLA3 also has retinyl-esterase activity and is involved in retinol metabolism. Expression of PNPLA3 gene and protein remain increased in activated HSCs. While induction of wild type of PNPLA3 is associated with reduced secretion of matrix metallopeptidase 2 and tissue inhibitor of metalloproteinase (1,2), HSCs with I148M show higher expression and release of proinflammatory cytokines (67-69). Donati et al. showed that a different PNPLA3 variant, rs2294918 E434K decreased the effect of the I148M variant (70). Lindén et al. studied antisense oligonucleotide (ASO) mediated silencing of PNPLA3 in a knock-in mice model. The ASO mediated silencing of PNPLA3 led to the reduction of hepatic steatosis, inflammation, NAFLD activity score and fibrosis stage (71). This study provides the first evidence that a PNPLA3 ASO therapy can improve all features of NAFLD including fibrosis.
Studying the ancestry-related and inter-individual differences in hepatic fat content and susceptibility to NAFLD, Romeo et al. conducted a genome-wide association scan of nonsynonymous sequence variations in Hispanics (n=383), African-Americans (n=1,032) and European-Americans (n=696). The authors found an allele of PNPLA3 [rs738409 (G), encoding I148M] to be strongly associated with increased hepatic fat levels and inflammation. This allele was most common in Hispanics, which were most susceptible to NAFLD. The hepatic fat content was more than two times higher in PNPLA3 rs738409 [G] homozygotes than in non-carriers. Another allele of PNPLA3 [rs6006460 (T), encoding S453I] was associated with lower hepatic fat content in African-Americans (26). Palmer et al. studied variants associated with NAFLD in persons with European ancestry, Africans and Hispanics. The authors found that steatosis was 0.20–0.34 heritable in Africans and Hispanic-American families. The variants in or near PNPLA3, neurocan (NCAN, rs2228603), glucokinase regulator (GCKR; rs780094), protein phosphatase 1, regulatory subunit 3B (PPP1R3B, rs4240624) were significantly associated with NAFLD in African Americans while PNPLA3 and PPP1R3B were significantly associated with NAFLD in Hispanic Americans (72). Table 1 (26-35) shows the GWAS studies in NAFLD. Rotman et al. analyzed data of 1,117 (894 adults and 223 children) individuals with histologically confirmed NAFLD. After adjustment for age, sex, diabetes mellitus (DM) and alcohol consumption, rs738409 C/G (PNPLA3-I148M) was associated with steatosis, portal inflammation, lobular inflammation, NAFLD activity score and fibrosis. Three SNPs on chromosome 10 near CHUK gene were independently associated with fibrosis. In children, no SNP was associated with histological severity (28). A meta-analysis of 16 studies showed that PNPLA3 rs738409 GG homozygous had 73% higher fat content when compared with CC polymorphism. Also, GG homozygous had greater risk of higher necroinflammatory scores and greater risk of developing fibrosis when compared with CC homozygous (3.24- and 3.2-fold respectively). NASH was more common in GG than CC homozygous [odds ratio (OR) 3.488; 95% CI, 1.859–6.545, data from 2,124 patients) (73).
Genetic variation in PNPLA3 is also associated with risk of HCC in NAFLD. Burza et al. showed for the first time, that PNPLA3 I148M allele is associated with risk of HCC. The authors compared bariatric surgery group to control group (conventional treatment for obesity) and found significantly higher incidence of HCC in the PNPLA3 I148M variant group in comparison to the control group (log-rank P value =0.001) (74). Liu et al. compared PNPLA3 rs738409 genotype in 100 European Caucasians with NAFLD related HCC to 275 controls with histological NAFLD. The multivariate analysis adjusted for age, gender, DM, BMI, and cirrhosis found that additive risk for HCC was 2.26 OR for each copy of gene. The GG homozygotes exhibited a 5-fold [1.47–17.29], P=0.01, increased risk over CC (75). Singal et al. performed a meta-analysis of 24 studies, which included 9,915 patients. PNPLA3 was associated with severity of fibrosis, OR being 1.32 (95% CI, 1.20–1.45). PNPLA3 increased fibrosis risk across all etiologies. Nine studies (n=2,937) showed increased risk of HCC in patients with cirrhosis (OR 1.40; 95% CI, 1.12–1.75), related to NASH or ALD, and not in other etiologies (76).
Transmembrane 6 superfamily 2 (TM6SF2) is a gene on chromosome 19p12, which functions as a lipid transporter. It is expressed predominantly in liver and intestine. The TM6SF2 encodes a protein with transmembrane domains. TM6SF2 is localized in the endoplasmic reticulum and ER-Golgi intermediate compartment of human liver cells. Functional studies were done in human hepatoma Huh7 and HepG2 cells, using siRNA inhibition and overexpression techniques. TM6SF2 siRNA inhibition was associated with reduced secretion of triglyceride rich lipoproteins and increased cellular triglyceride concentration and lipid droplet content, while TM6SF2 over expression reduced steatosis (77-79). TM6SF2 activity is required for normal VLDL secretion and impaired TM6SF2 function causally contributes to NAFLD (32). The Glu167Lys missense mutation was shown to alter serum lipid profiles in humans and the knockdown of TM6SF2 in mice was shown to have increase liver triglyceride content and decreased VLDL secretion (79). Prill et al. studied in vitro disease model based on 3D spheroids from human hepatocytes and found that the TM6SF2E167K variant increased hepatocyte fat content by reducing APOB particle secretion (80).
As TM6SF2 controls hepatic lipid efflux, decreased effect by deletions or mutations reduces secretion of lipoprotein, causing hepatocellular triglyceride accumulation (81). TM6SF2 rs58542926 T-allele mediated hepatic retention of triglycerides and cholesterol predispose to NAFLD related fibrosis, whereas C-allele carriage promotes VLDL excretion, thus increasing risk of CVD or atherosclerosis while protecting the liver (82). In an exome-wide association study, Kozlitina et al. identified a TM6SF2 variant that conferred susceptibility to nonalcoholic fatty liver disease (34). Dongiovanni et al. confirmed that patients with TM6SF2 E167K variant have severe fatty liver disease (more steatosis, inflammation, ballooning and fibrosis) and lower circulating lipids (thus having reduced risk of myocardial infarction). E167K carriers had higher ALT and lower lipid levels (P<0.05), as well as a lower incidence of cardiovascular events (41). Liu et al. studied two histological NAFLD cohorts consisting of steatosis, steatohepatitis, fibrosis and cirrhosis (n=1,074). The authors found significant association of TM6SF2 to advanced fibrosis/cirrhosis. This association was independent of age, BMI, T2DM and PNPLA3 rs738409 genotype (42). In contrast to association of PNPLA3 to HCC; there is less data on TM6SF2 and HCC risk. A recent study by Yang et al. included 1,020 HCC, 2,021 controls with chronic liver disease and 2,484 healthy individuals in discover cohort and 249 alcoholic cirrhosis and 268 hepatitis C cirrhosis in prospective cohort. The authors found significant association of PNPLA3 and TM6SF2 to risk of HCC. PNPLA3 SNP was also significantly associated with HCC in non-fibrotic liver (OR =2.19; 95% CI, 1.22–3.92, P=0.007) (83).
Other genetic modifiers in NAFLD
The Table 2 shows some of the candidate gene studies in NAFLD. The genetic associations based on candidate gene studies can be either specific to NAFLD (related to lipid metabolism, insulin resistance and glucose metabolism) or non-specific related to inflammation, oxidative stress and fibrosis. ENPP1 (ectoenzyme nucleotide pyrophosphate phosphodiesterase 1) and IRS-1 (insulin receptor substrate-1) polymorphisms affect insulin sensitivity. Dongiovanni et al. compared 702 patients with biopsy-proven NAFLD and 310 controls. The ENPP1-121Gln and 972Arg-IRS-1 polymorphisms were independently associated with fibrosis in multivariate analysis. Both polymorphisms were associated with a marked reduction of AKT activation status, reflecting insulin resistance in obese patients with NAFLD (36). Glucokinase regulatory protein (GCKR) is associated with lipid and glucose metabolism and GCKR C > T SNP (OR 2.06) has been shown to be independently associated with significant liver fibrosis (37). A study observed that variants of solute carrier family 2 [(facilitated glucose transporter) member 1] (SLC2A1) were associated with NAFLD, and in vitro down-regulation SLC2A1 promoted lipid accumulation and oxidative stress (38). The transcription factor 7-like 2 (TCF7L2) polymorphism predisposes to diabetes by modulating beta-cell function and modulates lipid levels in familial dyslipidemia. Musso et al. showed that TCF7L2 polymorphism predisposed to NAFLD and significantly impacted liver injury, glucose homeostasis, postprandial lipoprotein and adipokine responses to fat ingestion (39). Membrane bound O-acyltransferase domain- containing 7 (MBOAT7), which is also known as LPIAT1, is a protein involved in the acyl chain remodeling of phospholipids. Recently, it has been shown that MBOAT7 is a multispanning transmembrane protein with six transmembrane domains (84). MBOAT7 catalyzes transfer of polyunsaturated fatty acids, thus maintaining fluidity of cell membranes. MBOAT7 is involved in the re-acylation of phospholipids. The rs641738 gene variant leads to a reduced MBOAT7 expression favoring increase in free arachidonic acid, which is a driver of hepatic inflammation (85). Mancina et al. showed that rs641738 C > T variant of MBOAT7 increased hepatic fat content, severity and fibrosis in comparison to subjects without the variant (44).
Ma et al. analyzed role of the SNP rs6834314 and its nearest gene, 17-beta hydroxysteroid dehydrogenase 13 (HSD17B13) in 768 adult Caucasians patients with NAFLD. The enzyme is lipid droplet-associated retinol dehydrogenase. The minor allele of rs6834314 was significantly associated with increased steatosis, but decreased severity (inflammation, ballooning, Mallory-Denk bodies) and liver enzyme levels (52).
The fatty acid transport protein 5 (FATP5) is involved in hepatic lipid and bile metabolism. Oxidative stress plays an important role in pathogenesis of NASH and genes affecting oxidative stress have been shown to be associated with NASH. GCLC is involved in glutathione synthesis and variant is associated with steatohepatitis as compared to steatosis (55). Manganese-dependent superoxide dismutase (MnSOD) plays a role in protecting cells from oxidative stress. Al-Serri et al. showed that SOD2 C47T polymorphism was associated with more fibrosis in NASH (56). Uncoupling protein 2 (UCP2) is involved in mitochondrial lipid fluxes and reactive oxygen species production by the respiratory chain and it was observed that UCP2-866 A/A genotype was associated with increased hepatic UCP2 expression and reduced risk of NASH, particularly in subjects with normal glucose (57). Genes associated with inflammation also modify susceptibility of NAFLD/NASH. Valenti et al. analyzed 99 patients with NAFLD, 238 TNF-alpha polymorphism was higher in patients with NAFLD than in controls (31% vs. 15%; P=0.0001), and also these patients had had higher insulin resistance indices (58).
Iron overload is thought to be associated with severe forms of NAFLD; however, not all studies have found an association with HFE gene mutations (86-88). Apolipoproteins are proteins that bind to lipids to form lipoproteins, thus helping in transport of lipids and also have affinity to some receptors. While some studies have found an association of Apolipoprotein C3 gene variants with NAFLD, other studies have not shown an association (89,90).
Genetic basis in the pathogenesis of ALD
As stated earlier, severity of liver disease differs among patients consuming similar type and quantity of alcohol over a similar duration. Even though alcoholic steatosis develops in majority, most patients consuming unsafe amount of alcohol escape from liver injury and do not develop significant liver disease (severe alcoholic hepatitis or cirrhosis). In fact, of all patients who consume alcohol in significant amount, cirrhosis develops only in 8–20% of patients (5,91,92). Bellentani et al. analyzed data of 6,534 subjects and observed that the risk of developing cirrhosis increased with increasing daily intake, drinking alcohol outside mealtimes and with use of different alcoholic beverages (91). Environmental factors other than heavy alcohol consumption which affect progression of ALD include gender (females are more susceptible), obesity and coexistence of viral hepatitis but are still not able to explain the significant inter-individual differences in ALD (3). Thus, genetic and epigenetic factors may play an important role in the pathogenesis of ALD.
In a study comparing 580 cases and 279 controls (defined as free of significant liver disease despite similar alcohol consumption), cases were significantly more likely to report death of father due to ALD (odds ratio 2.53) (93). Reed et al. analyzed medical records of 15,924 twin-pairs from twin registry. The monozygotic twins had higher concordance of alcoholism (26.7 vs. 12.2 for dizygotic) and cirrhosis in comparison to dizygotic twins (6.9 vs. 5.3 for dizygotic, P<0.001) (11). In a meta-analysis of 12 twin and five adoption studies, heritability of alcohol use disorder was almost 50% (9). Analysing the inter-ethnic differences, Stinson et al. reported higher risk for alcoholic cirrhosis related mortality in Hispanics in comparison to African Americans non-Hispanics and white non-Hispanics (10).
Genetic modifiers in ALD
Multiple studies have shown PNPLA3 gene polymorphisms to be associated with ALD. Tian et al. showed that rs738409 variant of PNPLA3 was strongly associated with ALD (unadjusted OR =2.25) (94). Stickel et al. studied impact of rs738409 gene variant on manifestation of ALD in two German cohorts that included 1,043 alcoholic patients with or without ALD and in 376 at-risk drinkers. The rs738409 genotype GG was strongly over represented in patients with cirrhosis (OR 2.79) and in alcoholic patients with elevated ALT levels (OR 2.33). The population attributable risk of cirrhosis in carriers of PNPLA3 rs738409 (G) was estimated to be 26.6% (95). Several studies have shown that PNPLA3 is a risk factor for HCC in ALD (96-98). A study observed that patients with cirrhosis and HCC were more likely to be G/G homozygotes, and this happened more commonly in patients with alcohol/metabolic cirrhosis as compared to viral cirrhosis (96). Friedrich et al. studied I148M polymorphism in 421 Caucasian patients. The G allele of the I148M variant was significantly more common in patients with ALD and HCC. Also, transplant free survival was lower in these patients (97).
Data regarding the association of TM6SF2 gene variant with ALD is less robust in comparison to NAFLD. Buch et al. performed a GWAS for alcohol related cirrhosis in 712 cases and 1,426 controls; the authors also validated results in two independent European cohorts (1,148 cases and 922 controls) and found that membrane bound O-acyltransferase domain containing 7 (MBOAT7), TM6SF2 and PNPLA3 gene variants were associated with alcohol related cirrhosis. As all these genes are involved in lipid metabolism, it appears that lipid turnover is also important in the pathogenesis of alcohol related cirrhosis (99). Mancina et al. studied 416 at-risk alcohol drinkers retrospectively. The authors observed that PNPLA3, CD14 and TM6SF2 were associated with prevalence of alcoholic cirrhosis but only PNPLA3 and CD14 (and not TM6SF2) were associated with incidence of alcoholic cirrhosis (100). Further, in an Eastern European population, TM6SF2 rs58542926 and MBOAT7 rs641738 were not found to be related to alcohol related cirrhosis (101).
The other genetic polymorphisms shown to be associated with ALD are shown in Table 3 (94-110) and include alcohol metabolizing genes like alcohol dehydrogenase (ADH), aldehyde dehydrogenase (ALDH) and Cytochrome P450 2E1 (CYP2E1). ADH oxidizes alcohol to acetaldehyde, and acetaldehyde is further oxidized to acetate by ALDH. Both these steps require NAD+ as cofactor and both these reactions lead to a reduced NAD+/NADH ratio, which favors fatty acid synthesis and fat accumulation. CYP2E1 catalyzes ethanol oxidation to acetaldehyde, generating significant amount of reactive oxygen species, oxidative stress and inflammation. Formation of acetaldehyde and oxidative stress inhibit Peroxisome Proliferator Activated Receptor (PPAR) alpha transcriptional activity, decreasing fatty acid oxidation (85). The genetic variants of ADH, ALDH and CYP2E1 have also been shown to be associated alcoholic cirrhosis (102-106). The effect of CYP2E1 variants increases in the presence of other genetic variants involved in detoxification of reactive oxygen species (105,106). Genes involved in inflammation like cytotoxic T-lymphocyte associated protein 4, tumor necrosis factor and interleukin 1 beta are also associated with higher risk of cirrhosis (108-110). Figure 1 shows genetic modifiers of NAFLD and ALD.
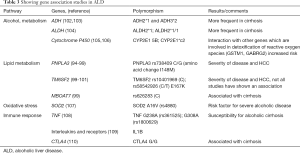
Full table
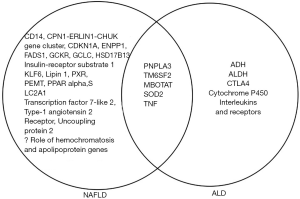
Epigenetic modifiers in NAFLD and ALD
Epigenetics include a process that alters gene activity without changing the DNA sequence. These changes can be transmitted to daughter cells by cell division. Epigenetic modifications are caused by alterations in DNA methylation, modifications in histone proteins and by micro RNAs (miR) (111-113). Epigenetic changes have been shown to affect both NAFLD (114) and ALD (85); however, knowledge and data regarding epigenetic changes is limited at present. miRNAs are transcribed in cell nucleus and transported to the cytoplasm, where they are processed into mature miRNAs. miRNAs are 19-22 nucleotide non-coding sequences that bind to the complementary sequence of messenger RNA molecules and regulate gene expression by silencing or inhibition of translation. miRNAs play a role in many cellular processes. miRNA dysregulation has been shown to be associated with several liver diseases including ALD, NAFLD, viral hepatitis, fibrosis and HCC (111,112). miRNAs remain stable in the circulation (blood and urine) and thus represent potential biomarkers for diseases (115). Following miRNAs are shown to be up regulated in ALD (in humans or animal models); miR-155, miR-34a, miR-212, miR-21, miR-181a, miR-217, miR-223. Following miRNAs are down regulated in ALD; miR-122, miR-29, miR-199a, miR-125b, miR-126, mi200a, miR-375. These miRNAs are involved in increased intestinal permeability, liver injury, inflammation, steatosis, oxidative stress, fibrosis, cirrhosis and HCC, in humans or animal models (85,116-118). Data in NAFLD patients suggests that following miRNAs are upregulated in NAFLD: miR-21, miR-34a, miR-182 whereas miR-122 is down regulated (114). Other epigenetic mechanisms which affect NAFLD and ALD include DNA methylation. In a study of DNA methylation of 4 CpG islands (CpG99, CpG71, CpG26 and CpG101) in regulatory regions of PNPLA3, SAMM50, PARVB variant 1, and PARVB variant 2, Kitamoto et al. showed hypomethylation of CpG26 (PARVB variant 1) and hypermethylation of CpG99 in the regulatory region of PNPLA3 which was associated with fibrosis in NAFLD (119). Cordero et al. showed that liver fat accumulation induced by a high-fat-sucrose diet in male Wistar rats was prevented by methyl donor supplementation (120). Zeybel et al. showed that differential DNA methylation at specific CpGs within genes affecting fibrogenesis distinguished mild from severe fibrosis in both NAFLD and ALD (121). Hardy et al. compared biopsy proven NAFLD patients with controls; differential DNA methylation at PPARγ promoter was detected within the pool of cell-free DNA of plasma. Similar changes were present in patients with alcoholic cirrhosis (122).
Modifications of the histone proteins are other epigenetic mechanism involved in the pathogenesis of NAFLD and ALD. The histones are involved in maintenance of chromatin structure and gene expression. Modifications described are the acetylation which causes activation of gene transcription and deacetylation causing repression of genes. The imbalance between enzymes causing acetylation and deacetylation may influence the phenotypic gene expression (123). The aberrant histone modifications promote development of insulin resistance and diabetes mellitus (124). cAMP responsive element-binding protein (CREBH) is a hepatocyte specific transcription factor, which is localized in the endoplasmic reticulum (ER) membrane. CREBH is activated by ER stress or inflammation, which enters into the cell nucleus and activates expression of genes involved in acute-phase response, gluconeogenesis, lipogenesis, fatty acid oxidation, and lipolysis. Thus, modulation of CREBH acetylation can lead to altered lipid homeostasis associated with NAFLD (123,125). The activation of deacetylase sirtuin-1 has potential against the physiological mechanisms related to NAFLD (126). Alcohol also has been shown to affect acetylation and phosphorylation of histones in rat hepatocytes (127).
In an example of epigenetic change transmitted to offspring, Bruce et al. showed that maternal fat intake during gestation in female mice contributed to development of NAFLD in offspring (128). Female mice were fed with either a high-fat (HF) or control chow (C) diet before and during gestation, and during lactation. The offspring were fed either a C or a HF diet after weaning, thus generating four offspring groups: HF/HF, HF/C, C/HF, C/C. The liver histology was normal in C/C and HF/C offspring at 15 weeks. The C/HF offspring developed NAFL while HF/HF offspring developed NASH. Histological analysis at 30 weeks showed NAFLD in HF/C and C/HF groups, whereas the HF/HF had a more severe form of NASH. Thus, exposure to HF diet in utero and during lactation contributed to severe form of disease. Hepatic mitochondrial electron transport chain enzyme complex activity was reduced in offspring from HF-fed mothers. Also, lipogenesis, oxidative stress, and inflammatory pathways were up regulated at 15 weeks in offsprings of HF mothers (128).
Clinical implications of genetic and epigenetic modifiers in NAFLD and ALD
Genetic information can change occurrence of a disease in several ways. First, it may lead to behaviour change of a subject, thus decreasing risk of disease. Second, disease can be diagnosed timely or can be prevented by timely intervention. Third, pharmacotherapy can be tailored as per genetic information. As fibrosis progression in both NAFLD and ALD spans many years, a timely diagnosis/intervention of at risk subjects can prevent development of cirrhosis. As PNPLA3 is associated with higher risk of HCC, the knowledge of genetic modifiers may also help in identifying patients who would need strict HCC surveillance (75). A lower threshold may be kept in patients with TM6SF2 variants for cardiovascular disease screening (82). Genetic and epigenetic modifiers may alter response to pharmacotherapy and thus may help in modifying the treatment options. Dongiovanni et al. looked at response to statins in 107 patients with NAFLD. Use of statins was significantly associated with protection from steatosis, NASH, and fibrosis, but this effect was stronger in patients without I148M PNPLA3 risk variant. Thus, I148M PNPLA3 variant limited efficacy of statins (129). Scorletti et al. studied 103 patients with NAFLD, randomized to omega-3 fatty acids (DHA + EPA) or placebo for 15–18 months. Fifty-five men and 40 women completed the study. PNPLA3 148M/M variant influenced the changes in liver fat and DHA tissue enrichment (130). Patients with PNPLA3 polymorphism have been shown to respond better to life style modifications (131). Since pathogenesis of NAFLD is complex and is affected by several environmental and genetic factors, a single genetic variant is unlikely to have very strong role in risk prediction; however, a combination score of several genetic variants may provide useful insight into disease progression in future. Whether genetic information can modify disease risk of NAFLD or ALD, is yet to be seen. How genetic information can modify diet was shown by Nielsen et al. The authors conducted a double-blinded randomized controlled trial to examine the short- and long-term effects of genetic information on nutrition and dietary intake of caffeine, vitamin C, added sugars and sodium. The intervention group (n=92) received genetic information based dietary advice and the control group (n=46) received general dietary recommendations without genetic information for 12-month. At 12-month, the participants with a risk type of ACE gene in the intervention group significantly reduced sodium intake compared to the control group (132). The genetic information about risk of future development of NAFLD or ALD should lead to change of risk behavior and decrease of modifiable risk factors like diet, exercise, avoidance of weight gain or weight loss (if obese), decrease in amount/frequency of alcohol consumption.
Conclusions
A significant variability in disease severity of NAFLD and ALD occur due to genetic modifiers. Epigenetics changes may explain the phenotypic variability in patients with similar gene polymorphism. Knowledge of genetic and epigenetic modifiers should lead to development of new therapeutic targets and more selected therapy for patients with NAFLD and ALD. Also, better risk prediction regarding development of cirrhosis and HCC should be feasible in future with better understanding of these factors.
Acknowledgments
Mr. Yogesh Saini (Research Coordinator).
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Ashwani K. Singal) for the series “Non-alcoholic Fatty Liver Disease and Alcoholic Liver Disease” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh.2019.09.06). The series “Non-alcoholic Fatty Liver Disease and Alcoholic Liver Disease” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328-57. [Crossref] [PubMed]
- Duseja A, Singh SP, Saraswat VA, et al. Non-alcoholic fatty liver disease and metabolic syndrome-position paper of the Indian National Association for the study of the liver, endocrine society of India, Indian college of cardiology and Indian society of gastroenterology. J Clin Exp Hepatol 2015;5:51-68. [Crossref] [PubMed]
- Singal AK, Bataller R, Ahn J, et al. ACG Clinical Guideline: Alcoholic Liver Disease. Am J Gastroenterol 2018;113:175-94. [Crossref] [PubMed]
- Calzadilla Bertot L, Adams LA. The Natural Course of Non-Alcoholic Fatty Liver Disease. Int J Mol Sci 2016;17:774. [Crossref] [PubMed]
- Schwartz JM, Reinus JF. Prevalence and natural history of alcoholic liver disease. Clin Liver Dis 2012;16:659-66. [Crossref] [PubMed]
- Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology 2004;40:1387-95. [Crossref] [PubMed]
- Rich NE, Oji S, Mufti AR, et al. Racial and Ethnic Disparities in Nonalcoholic Fatty Liver Disease Prevalence, Severity, and Outcomes in the United States: A Systematic Review and Meta-analysis. Clin Gastroenterol Hepatol 2018;16:198-210.e2. [Crossref] [PubMed]
- Brouwers MC, Cantor RM, Kono N, et al. Heritability and genetic loci of fatty liver in familial combined hyperlipidemia. J Lipid Res 2006;47:2799-807. [Crossref] [PubMed]
- Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 2015;45:1061-72. [Crossref] [PubMed]
- Stinson FS, Grant BF, Dufour MC. The critical dimension of ethnicity in liver cirrhosis mortality statistics. Alcohol Clin Exp Res 2001;25:1181-7. [Crossref] [PubMed]
- Reed T, Page WF, Viken RJ, et al. Genetic predisposition to organ-specific endpoints of alcoholism. Alcohol Clin Exp Res 1996;20:1528-33. [Crossref] [PubMed]
- Singh S, Allen AM, Wang Z, et al. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol 2015;13:643-54.e1-9; quiz e39-40.
- McPherson S, Hardy T, Henderson E, et al. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J Hepatol 2015;62:1148-55. [Crossref] [PubMed]
- Struben VM, Hespenheide EE, Caldwell SH. Nonalcoholic steatohepatitis and cryptogenic cirrhosis within kindreds. Am J Med 2000;108:9-13. [Crossref] [PubMed]
- Schwimmer JB, Celedon MA, Lavine JE, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology 2009;136:1585-92. [Crossref] [PubMed]
- Makkonen J, Pietiläinen KH, Rissanen A, et al. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: a study in monozygotic and dizygotic twins. J Hepatol 2009;50:1035-42. [Crossref] [PubMed]
- Tarnoki AD, Tarnoki DL, Bata P, et al. Heritability of non-alcoholic fatty liver disease and association with abnormal vascular parameters: a twin study. Liver Int 2012;32:1287-93. [Crossref] [PubMed]
- Loomba R, Schork N, Chen CH, et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology 2015;149:1784-93. [Crossref] [PubMed]
- Cui J, Chen CH, Lo MT, et al. Shared genetic effects between hepatic steatosis and fibrosis: A prospective twin study. Hepatology 2016;64:1547-58. [Crossref] [PubMed]
- Browning JD, Kumar KS, Saboorian MH, et al. Ethnic differences in the prevalence of cryptogenic cirrhosis. Am J Gastroenterol 2004;99:292-8. [Crossref] [PubMed]
- Wagenknecht LE, Scherzinger AL, Stamm ER, et al. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring) 2009;17:1240-6. [PubMed]
- Bambha K, Belt P, Abraham M, et al. Ethnicity and nonalcoholic fatty liver disease. Hepatology 2012;55:769-80. [Crossref] [PubMed]
- Mohanty SR, Troy TN, Huo D, et al. Influence of ethnicity on histological differences in non-alcoholic fatty liver disease. J Hepatol 2009;50:797-804. [Crossref] [PubMed]
- Tabibian JH, Lazo M, Durazo FA, et al. Nonalcoholic fatty liver disease across ethno-racial groups: do Asian-American adults represent a new at-risk population? J Gastroenterol Hepatol 2011;26:501-9. [Crossref] [PubMed]
- Petersen KF, Dufour S, Feng J, et al. Increased prevalence of insulin resistance and nonalcoholic fatty liver disease in Asian-Indian men. Proc Natl Acad Sci U S A 2006;103:18273-7. [Crossref] [PubMed]
- Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2008;40:1461-5. [Crossref] [PubMed]
- Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 2008;83:520-8. [Crossref] [PubMed]
- Rotman Y, Koh C, Zmuda JM, et al. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology 2010;52:894-903. [Crossref] [PubMed]
- Chalasani N, Guo X, Loomba R, et al. Genome-wide association study identifies variants associated with histologic features of nonalcoholic Fatty liver disease. Gastroenterology 2010;139:1567-76. [Crossref] [PubMed]
- Chambers JC, Zhang W, Sehmi J, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet 2011;43:1131-8. [Crossref] [PubMed]
- Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 2011;7:e1001324. [Crossref] [PubMed]
- Kawaguchi T, Sumida Y, Umemura A, et al. Genetic Polymorphisms of the Human PNPLA3 Gene Are Strongly Associated with Severity of Non-Alcoholic Fatty Liver Disease in Japanese. PLoS One 2012;7:e38322. [Crossref] [PubMed]
- Feitosa MF, Wojczynski MK, North KE, et al. The ERLIN1-CHUK-CWF19L1 gene cluster influences liver fat deposition and hepatic inflammation in the NHLBI. Family Heart Study. Atherosclerosis 2013;228:175-80. [Crossref] [PubMed]
- Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 2014;46:352-6. [Crossref] [PubMed]
- DiStefano JK, Kingsley C, Craig Wood G, et al. Genomewide analysis of hepatic lipid content in extreme obesity. Acta Diabetol 2015;52:373-82. [Crossref] [PubMed]
- Dongiovanni P, Valenti L, Rametta R, et al. Genetic variants regulating insulin receptor signalling are associated with the severity of liver damage in patients with non-alcoholic fatty liver disease. Gut 2010;59:267-73. [Crossref] [PubMed]
- Petta S, Miele L, Bugianesi E, et al. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS One 2014;9:e87523. [Crossref] [PubMed]
- Vazquez-Chantada M, Gonzalez-Lahera A, Martinez-Arranz I, et al. Solute carrier family 2 member 1 is involved in the development of nonalcoholic fatty liver disease. Hepatology 2013;57:505-14. [Crossref] [PubMed]
- Musso G, Gambino R, Pacini G, et al. Transcription factor 7-like 2 polymorphism modulates glucose and lipid homeostasis, adipokine profile, and hepatocyte apoptosis in NASH. Hepatology 2009;49:426-35. [Crossref] [PubMed]
- Krawczyk M, Rau M, Schattenberg JM, et al. Combined effects of the PNPLA3 rs738409, TM6SF2 rs58542926, and MBOAT7 rs641738 variants on NAFLD severity: a multicenter biopsy-based study. J Lipid Res 2017;58:247-55. [Crossref] [PubMed]
- Dongiovanni P, Petta S, Maglio C, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology 2015;61:506-14. [Crossref] [PubMed]
- Liu YL, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun 2014;5:4309. [Crossref] [PubMed]
- Li Y, Liu S, Gao Y, et al. Association of TM6SF2 rs58542926 gene polymorphism with the risk of non-alcoholic fatty liver disease and colorectal adenoma in Chinese Han population. BMC Biochem 2019;20:3. [Crossref] [PubMed]
- Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016;150:1219-30.e6. [Crossref] [PubMed]
- Wang L, Athinarayanan S, Jiang G, et al. Fatty acid desaturase 1 gene polymorphisms control human hepatic lipid composition. Hepatology 2015;61:119-28. [Crossref] [PubMed]
- Valenti L, Motta BM, Alisi A, et al. LPIN1 rs13412852 polymorphism in pediatric nonalcoholic fatty liver disease. J Pediatr Gastroenterol Nutr 2012;54:588-93. [Crossref] [PubMed]
- Sookoian S, Castano GO, Burgueno AL, et al. The nuclear receptor PXR gene variants are associated with liver injury in nonalcoholic fatty liver disease. Pharmacogenet Genomics 2010;20:1-8. [Crossref] [PubMed]
- Chen S, Li Y, Li S, et al. A Val227Ala substitution in the peroxisome proliferator activated receptor alpha (PPAR alpha) gene associated with non-alcoholic fatty liver disease and decreased waist circumference and waist-to-hip ratio. J Gastroenterol Hepatol 2008;23:1415-8. [Crossref] [PubMed]
- Yamakawa-Kobayashi K, Ishiguro H, Arinami T, et al. A Val227Ala polymorphism in the peroxisome proliferator activated receptor alpha (PPARalpha) gene is associated with variations in serum lipid levels. J Med Genet 2002;39:189-91. [Crossref] [PubMed]
- Dong H, Wang J, Li C, et al. The phosphatidylethanolamine N-methyltransferase gene V175M single nucleotide polymorphism confers the susceptibility to NASH in Japanese population. J Hepatol 2007;46:915-20. [Crossref] [PubMed]
- Song J, da Costa KA, Fischer LM, et al. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD). FASEB J 2005;19:1266-71. [Crossref] [PubMed]
- Ma Y, Belyaeva OV, Brown PM, et al. 17-Beta Hydroxysteroid Dehydrogenase 13 Is a Hepatic Retinol Dehydrogenase Associated With Histological Features of Nonalcoholic Fatty Liver Disease. Hepatology 2019;69:1504-19. [Crossref] [PubMed]
- Yoneda M, Hotta K, Nozaki Y, et al. Association between angiotensin II type 1 receptor polymorphisms and the occurrence of nonalcoholic fatty liver disease. Liver Int 2009;29:1078-85. [Crossref] [PubMed]
- Zain SM, Mohamed Z, Mahadeva S, et al. Susceptibility and gene interaction study of the angiotensin II type 1 receptor (AGTR1) gene polymorphisms with non-alcoholic fatty liver disease in a multi-ethnic population. PLoS One 2013;8:e58538. [Crossref] [PubMed]
- Oliveira CP, Stefano JT, Cavaleiro AM, et al. Association of polymorphisms of glutamate-cystein ligase and microsomal triglyceride transfer protein genes in non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2010;25:357-61. [Crossref] [PubMed]
- Al-Serri A, Anstee QM, Valenti L, et al. The SOD2 C47T polymorphism influences NAFLD fibrosis severity: evidence from case-control and intra-familial allele association studies. J Hepatol 2012;56:448-54. [Crossref] [PubMed]
- Fares R, Petta S, Lombardi R, et al. The UCP2 -866 G>A promoter region polymorphism is associated with nonalcoholic steatohepatitis. Liver Int 2015;35:1574-80. [Crossref] [PubMed]
- Valenti L, Fracanzani A L, Dongiovanni P, et al. Tumor necrosis factor alpha promoter polymorphisms and insulin resistance in nonalcoholic fatty liver disease. Gastroenterology 2002;122:274-80. [Crossref] [PubMed]
- Tokushige K, Takakura M, Tsuchiya-Matsushita N, et al. Influence of TNF gene polymorphisms in Japanese patients with NASH and simple steatosis. J Hepatol 2007;46:1104-10. [Crossref] [PubMed]
- Kapil S, Duseja A, Sharma BK, et al. Genetic polymorphism in CD14 gene, a co-receptor of TLR4 associated with non-alcoholic fatty liver disease. World J Gastroenterol 2016;22:9346-55. [Crossref] [PubMed]
- Aravinthan A, Mells G, Allison M, et al. Gene polymorphisms of cellular senescence marker p21 and disease progression in non-alcohol-related fatty liver disease. Cell Cycle 2014;13:1489-94. [Crossref] [PubMed]
- Miele L, Beale G, Patman G, et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology 2008;135:282-91.e1. [Crossref] [PubMed]
- Anstee QM, Seth D, Day CP, et al. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology 2016;150:1728-44.e7. [Crossref] [PubMed]
- He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 2010;285:6706-15. [Crossref] [PubMed]
- Pirazzi C, Adiels M, Burza MA, et al. Patatin-like phospholipase domain-containing 3 (PNPLA3) I148M (rs738409) affects hepatic VLDL secretion in humans and in vitro. J Hepatol 2012;57:1276-82. [Crossref] [PubMed]
- Kumari M, Schoiswohl G, Chitraju C, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab 2012;15:691-702. [Crossref] [PubMed]
- Pirazzi C, Valenti L, Motta BM, et al. PNPLA3 has retinyl-palmitate lipase activity in human hepatic stellate cells. Hum Mol Genet 2014;23:4077-85. [Crossref] [PubMed]
- Pingitore P, Dongiovanni P, Motta BM, et al. PNPLA3 overexpression results in reduction of proteins predisposing to fibrosis. Hum Mol Genet 2016;25:5212-22. [PubMed]
- Bruschi FV, Claudel T, Tardelli M, et al. The PNPLA3 I148M variant modulates the fibrogenic phenotype of human hepaticstellate cells. Hepatology 2017;65:1875-90. [Crossref] [PubMed]
- Donati B, Motta BM, Pingitore P, et al. The rs2294918 E434K variant modulates patatin-like phospholipase domain-containing 3 expression and liver damage. Hepatology 2016;63:787-98. [Crossref] [PubMed]
- Lindén D, Ahnmark A, Pingitore P, et al. Pnpla3 silencing with antisense oligonucleotides ameliorates nonalcoholic steatohepatitis and fibrosis in Pnpla3 I148M knock-in mice. Mol Metab 2019;22:49-61. [Crossref] [PubMed]
- Palmer ND, Musani SK, Yerges-Armstrong LM, et al. Characterization of European ancestry nonalcoholic fatty liver disease-associated variants in individuals of African and Hispanic descent. Hepatology 2013;58:966-75. [Crossref] [PubMed]
- Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 2011;53:1883-94. [Crossref] [PubMed]
- Burza MA, Pirazzi C, Maglio C, et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis 2012;44:1037-41. [Crossref] [PubMed]
- Liu YL, Patman GL, Leathart JB, et al. Carriage of the PNPLA3 rs738409 C>G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol 2014;61:75-81. [Crossref] [PubMed]
- Singal AG, Manjunath H, Yopp AC, et al. The effect of PNPLA3 on fibrosis progression and development of hepatocellular carcinoma: a meta-analysis. Am J Gastroenterol 2014;109:325-34. [Crossref] [PubMed]
- Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A 2014;111:8913-8. [Crossref] [PubMed]
- Carim-Todd L, Escarceller M, Estivill X, et al. Cloning of the novel gene TM6SF1 reveals conservation of clusters of paralogous genes between human chromosomes 15q24-->q26 and 19p13.3-->p12. Cytogenet Cell Genet 2000;90:255-60. [Crossref] [PubMed]
- Surakka I, Horikoshi M, Magi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet 2015;47:589-97. [Crossref] [PubMed]
- Prill S, Caddeo A, Baselli G, et al. The TM6SF2 E167K genetic variant induces lipid biosynthesis and reduces apolipoprotein B secretion in human hepatic 3D spheroids. Sci Rep 2019;9:11585. [Crossref] [PubMed]
- Holmen OL, Zhang H, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet 2014;46:345-51. [Crossref] [PubMed]
- Kahali B, Liu YL, Daly AK, et al. TM6SF2: catch-22 in the fight against nonalcoholic fatty liver disease and cardiovascular disease? Gastroenterology 2015;148:679-84. [Crossref] [PubMed]
- Yang J, Trépo E, Nahon P, et al. PNPLA3 and TM6SF2 variants as risk factors of hepatocellular carcinoma across various etiologies and severity of underlying liver diseases. Int J Cancer 2019;144:533-44. [Crossref] [PubMed]
- Caddeo A, Jamialahmadi O, Solinas G, et al. MBOAT7 is anchored to endomembranes by six transmembrane domains. J Struct Biol 2019;206:349-60. [Crossref] [PubMed]
- Meroni M, Longo M, Rametta R, et al. Genetic and Epigenetic Modifiers of Alcoholic Liver Disease. Int J Mol Sci 2018;19:3857. [Crossref] [PubMed]
- Duseja A, Das R, Nanda M, et al. Nonalcoholic steatohepatitis in Asian Indians is neither associated with iron overload nor with HFE gene mutations. World J Gastroenterol 2005;11:393-5. [Crossref] [PubMed]
- Saremi L, Lotfipanah S, Mohammadi M, et al. Association of HFE gene mutations with nonalcoholic fatty liver disease in the Iranian population. Cell Mol Biol (Noisy-le-grand) 2016;62:123-8. [PubMed]
- Nelson JE, Bhattacharya R, Lindor KD, et al. HFE C282Y mutations are associated with advanced hepatic fibrosis in Caucasians with nonalcoholic steatohepatitis. Hepatology 2007;46:723-9. [Crossref] [PubMed]
- Petersen KF, Dufour S, Hariri A, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med 2010;362:1082-9. [Crossref] [PubMed]
- Duseja A, Aggarwal R. APOC3 and PNPLA3 in non-alcoholic fatty liver disease: need to clear the air. J Gastroenterol Hepatol 2012;27:848-51. [Crossref] [PubMed]
- Bellentani S, Saccoccio G, Costa G, et al. Drinking habits as cofactors of risk for alcohol induced liver damage. The Dionysos Study Group. Gut 1997;41:845-50. [Crossref] [PubMed]
- Becker U, Deis A, Sorensen TI, et al. Prediction of risk of liver disease by alcohol intake, sex, and age: a prospective population study. Hepatology 1996;23:1025-9. [Crossref] [PubMed]
- Whitfield JB, Rahman K, Haber PS, et al. Brief report: genetics of alcoholic cirrhosis-GenomALC multinational study. Alcohol Clin Exp Res 2015;39:836-42. [Crossref] [PubMed]
- Tian C, Stokowski RP, Kershenobich D, et al. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet 2010;42:21-3. [Crossref] [PubMed]
- Stickel F, Buch S, Lau K, et al. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology 2011;53:86-95. [Crossref] [PubMed]
- Falleti E, Fabris C, Cmet S, et al. PNPLA3 rs738409 C/G polymorphism in cirrhosis: Relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int 2011;31:1137-43. [Crossref] [PubMed]
- Friedrich K, Wannhoff A, Kattner S, et al. PNPLA3 in end-stage liver disease: Alcohol consumption, hepatocellular carcinoma development, and transplantation-free survival. J Gastroenterol Hepatol 2014;29:1477-84. [Crossref] [PubMed]
- Trépo E, Nahon P, Bontempi G, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology 2014;59:2170-7. [Crossref] [PubMed]
- Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat Genet 2015;47:1443-8. [Crossref] [PubMed]
- Mancina RM, Ferri F, Farcomeni A, et al. A two gene-based risk score predicts alcoholic cirrhosis development in males with at-risk alcohol consumption. Appl Clin Genet 2019;12:1-10. [Crossref] [PubMed]
- Basyte-Bacevice V, Skieceviciene J, Valantiene I, et al. TM6SF2 and MBOAT7 Gene Variants in Liver Fibrosis and Cirrhosis. Int J Mol Sci 2019;20:1277. [Crossref] [PubMed]
- Chao YC, Wang MF, Tang HS, et al. Genotyping of alcohol dehydrogenase at the ADH2 and ADH3 loci by using a polymerase chain reaction and restriction-fragment-length polymorphism in Chinese alcoholic cirrhotics and non-alcoholics. Proc Natl Sci Counc Repub China B 1994;18:101-6. [PubMed]
- Monzoni A, Masutti F, Saccoccio G, et al. Genetic determinants of ethanol-induced liver damage. Mol Med 2001;7:255-62. [Crossref] [PubMed]
- Tanaka F, Shiratori Y, Yokosuka O, et al. High incidence of ADH2*1/ALDH2*1 genes among Japanese alcohol dependents and patients with alcoholic liver disease. Hepatology 1996;23:234-9. [Crossref] [PubMed]
- Khan AJ, Ruwali M, Choudhuri G, et al. Polymorphism in cytochrome P450 2E1 and interaction with other genetic risk factors and susceptibility to alcoholic liver cirrhosis. Mutat Res 2009;664:55-63. [Crossref] [PubMed]
- García-Bañuelos J, Panduro A, Gordillo-Bastidas D, et al. Genetic polymorphisms of genes coding to alcohol-metabolizing enzymes in western Mexicans: association of CYP2E1*c2/CYP2E1*5B allele with cirrhosis and liver function. Alcohol Clin Exp Res 2012;36:425-31. [Crossref] [PubMed]
- Degoul F, Sutton A, Mansouri A, et al. Homozygosity for alanine in the mitochondrial targeting sequence of superoxide dismutase and risk for severe alcoholic liver disease. Gastroenterology 2001;120:1468-74. [Crossref] [PubMed]
- Marcos M, Gomez-Munuera M, Pastor I, et al. Tumor necrosis factor polymorphisms and alcoholic liver disease: a HuGE review and meta-analysis. Am J Epidemiol 2009;170:948-56. [Crossref] [PubMed]
- Takamatsu M, Yamauchi M, Maezawa Y, et al. Genetic polymorphisms of interleukin-1beta in association with the development of alcoholic liver disease in Japanese patients. Am J Gastroenterol 2000;95:1305-11. [PubMed]
- Valenti L, De Feo T, Fracanzani AL, et al. Cytotoxic T-lymphocyte antigen-4 A49G polymorphism is associated with susceptibility to and severity of alcoholic liver disease in Italian patients. Alcohol Alcohol 2004;39:276-80. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Bala S. Emerging role of microRNAs in liver diseases. World J Gastroenterol 2009;15:5633-40. [Crossref] [PubMed]
- Lakshman R, Shah R, Reyes-Gordillo K, et al. Synergy between NAFLD and AFLD and potential biomarkers. Clin Res Hepatol Gastroenterol 2015;39:S29-34. [Crossref] [PubMed]
- Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J Hepatol 2018;68:268-79. [Crossref] [PubMed]
- Starkey Lewis PJ, Dear J, Platt V, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 2011;54:1767-76. [Crossref] [PubMed]
- Bala S, Szabo G. MicroRNA signature in alcoholic liver disease. Int J Hepatol 2012;2012:498232.
- Bala S, Marcos M, Kodys K, et al. Up-regulation of microRNA-155 in macrophages contributes to increased Tumor Necrosis Factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011;286:1436-44. [Crossref] [PubMed]
- Tang Y, Banan A, Forsyth CB, et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcoholism 2008;32:355-64. [Crossref] [PubMed]
- Kitamoto T, Kitamoto A, Ogawa Y, et al. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol 2015;63:494-502. [Crossref] [PubMed]
- Cordero P, Campion J, Milagro FI, et al. Transcriptomic and epigenetic changes in early liver steatosis associated to obesity: effect of dietary methyl donor supplementation. Mol Genet Metab 2013;110:388-95. [Crossref] [PubMed]
- Zeybel M, Hardy T, Robinson SM, et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenetics 2015;7:25. [Crossref] [PubMed]
- Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: a potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut 2017;66:1321-8. [Crossref] [PubMed]
- Del Campo JA, Gallego-Durán R, Gallego P, et al. Genetic and Epigenetic Regulation in Nonalcoholic Fatty Liver Disease (NAFLD). Int J Mol Sci 2018. [Crossref] [PubMed]
- Ling C, Groop L. Epigenetics: A molecular link between environmental factors and type 2 diabetes. Diabetes 2009;58:2718-25. [Crossref] [PubMed]
- Kim H, Mendez R, Chen X, et al. Lysine Acetylation of CREBH Regulates Fasting-Induced Hepatic Lipid Metabolism. Mol Cell Biol 2015;35:4121-34. [Crossref] [PubMed]
- Mariani S, Fiore D, Basciani S, et al. Plasma levels of SIRT1 associate with non-alcoholic fatty liver disease in obese patients. Endocrine 2015;49:711-6. [Crossref] [PubMed]
- Mandrekar P. Epigenetic regulation in alcoholic liver disease. World J Gastroenterol 2011;17:2456-64. [Crossref] [PubMed]
- Bruce KD, Cagampang FR, Argenton M, et al. Maternal high-fat feeding primes steatohepatitis in adult mice offspring, involving mitochondrial dysfunction and altered lipogenesis gene expression. Hepatology 2009;50:1796-808. [Crossref] [PubMed]
- Dongiovanni P, Petta S, Mannisto V, et al. Statin use and non-alcoholic steatohepatitis in at risk individuals. J Hepatol 2015;63:705-12. [Crossref] [PubMed]
- Scorletti E, West AL, Bhatia L, et al. Treating liver fat and serum triglyceride levels in NAFLD, effects of PNPLA3 and TM6SF2 genotypes: results from the WELCOME trial. J Hepatol 2015;63:1476-83. [Crossref] [PubMed]
- Shen J, Wong GL, Chan HL, et al. PNPLA3 gene polymorphism and response to lifestyle modification in patients with nonalcoholic fatty liver disease. J Gastroenterol Hepatol 2015;30:139-46. [Crossref] [PubMed]
- Nielsen DE, El-Sohemy A. Disclosure of genetic information and change in dietary intake: a randomized controlled trial. PLoS One 2014;9:e112665. [Crossref] [PubMed]
Cite this article as: Choudhary NS, Duseja A. Genetic and epigenetic disease modifiers: non-alcoholic fatty liver disease (NAFLD) and alcoholic liver disease (ALD). Transl Gastroenterol Hepatol 2021;6:2.


