
Genome editing technologies to treat rare liver diseases
Introduction
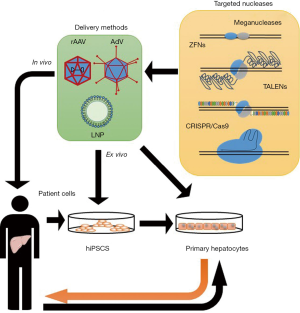
Rare liver diseases are a heterogeneous group of conditions that might affect different organs beside the liver, since hepatocytes have a large range of activities, particularly in lipid and protein metabolism and in detoxification: alterations involving liver cells lead to many different diseases, driving to the accumulation of proteins or toxic byproducts, with consequent damage for the whole organism. Liver transplantation, particularly in pediatric age, is the only resolutive therapy so far for most rare liver diseases, but it is limited by organ availability, post-operative complications and side-effects from immunosuppressive therapy (1). Most rare hereditary liver diseases are caused by loss-of-function mutations or diminished gene expression, and are thus promising candidates for both traditional and innovative gene therapy approaches: the main objective of traditional hepatic gene therapy has been to obtain an optimal delivery of the therapeutic gene within hepatocytes, in order to increase expression level of deficient protein and subsequently correcting the disease phenotype. Hepatocytes are easy to target, in particular with recombinant adeno-associated viruses (rAAV), which are already approved in clinical settings. As demonstrated in animal models and in humans, rAAV can be intravenously administered with adequate efficacy and may achieve prolonged transgene expression. Nevertheless, rAAV-based approaches have limitations such as vector immunogenicity and pre-existing immunity to AAV in humans (2). Lipid nanoparticles (LNPs) have also been employed for efficient delivery of transgenes in mouse liver, and showed the advantage to be biodegradable and well tolerated (3). Recently, the efforts to provide overexpression of defective gene products have shifted to direct correction of altered genomic DNA sequences.
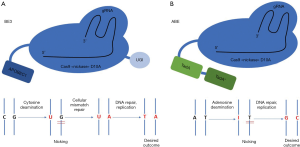
In the last years, many different genome editing platforms have been developed, such as meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeat (CRISPR)-associated nucleases. These techniques [whose main features and mechanisms of action have been widely reviewed (4,5) and are summarized in Table 1] have deeply revolutionized targeted genomic manipulation, opening unprecedented scenarios for both research and therapeutic applications, e.g., infectious diseases (6). The main feature of these approaches is the requirement for nucleases that generate double strand breaks (DSBs), that activate non–homologous end joining (NHEJ) as DNA repair process, often resulting in random insertions or deletions (indels) at the site of cleavage. If a homologous DNA template is present, homology-directed repair (HDR) can replace the DNA near to the cleavage site. Unfortunately, HDR is less active than NHEJ, and unwanted indels are frequently generated. Being HDR not highly efficient, particularly in non-dividing cells, the need for alternative approaches aimed at correcting point mutations and not relying on DSBs led researchers to investigate mechanisms for programmable direct conversion of one DNA base to another, in order to increase the efficiency of correction without introduction of unwanted random indels. Komor and colleagues engineered fusions of CRISPR/Cas9 and cytidine deaminase together with an RNA guide to obtain direct targeted conversion of cytidine to uridine, resulting in a C>T substitution without DSB. They obtained a first generation of cytosine base editors (BE1) that they subsequently improved by the addition of an enzyme capable of inhibiting cellular base excision repair process, thus obtaining BE2. In the so-called BE3, base editing efficiency was furtherly augmented by enhancing cell-mediated correction of the non-edited strand (7,8) (Figure 1). Since about half of pathogenic point mutations found in humans are caused by spontaneous deamination of cytosine, leading to transitions from C-G to T-A base pairs, adenine base editors (ABE) were also created (Figure 1). These engineered proteins converted targeted A-T to G-C base pairs with high efficiency, high product purity and low rate of indels (8,9). Together with CBEs, ABEs enable the targeted introduction of all four transition mutations with no need for DSBs, paving the way for a huge number of applications. Nevertheless, recent studies highlighted a remarkable occurrence of off-target effects of base editor tools. Jin et al. in rice (10) and Zuo et al. in mouse embryos (11) showed that, while ABEs cause rare unwanted substitutions, cytosine BE3 induce substantial genome-wide off-target single-nucleotide variants (SNVs), concluding that the fidelity of base editing platforms needs a significant improvement. Although these findings point out as base editors are still far from being considered for therapeutic applications, other editing techniques are considerably near to clinics. In this Review we provide an overview on different approaches for rare genetic liver diseases (summarized in Figure 2), such as metabolic disorders and hemophilia that have been explored so far as candidate for genome editing-based therapeutics. Hypercholesterolemia was also included in the discussion given the large number of genome editing approaches focusing on this disease. Moreover, a special consideration has been given to the literature reporting the use of base editors technology.

Full table
Phenylketonuria (PKU)
PKU is an autosomal recessive inborn error of hepatocyte metabolism caused by mutations in the PAH gene, encoding for the phenylalanine hydroxylase enzyme (PAH). PAH acts in phenylalanine catabolism pathway by catalyzing the hydroxylation of phenylalanine in tyrosine, and loss in its activity results in a raise of blood phenylalanine level, that eventually lead to a neurotoxic effect. PKU, if untreated, is associated with impaired postnatal cognitive development, often accompanied by other symptoms such as seizures, autism, motor deficits, eczematous rashes (12). The main treatment for PKU remains dietary restriction for phenylalanine that is unfortunately very difficult to achieve and encounters poor compliance from patients. Other potential therapeutic strategies include the use of competitors of phenylalanine uptake at enteric and blood-brain barrier level, or treatment with tetrahydrobiopterin for enzyme enhancement therapy (13). The ideal approach to reach PKU cure is the restoration of PAH activity, that might be achieved by the correction of pathogenic variants in PAH gene. Since PKU has autosomal recessive inheritance, patients carry PAH mutations on both alleles: nevertheless, the correction of only one allele could enable a raise in PAH enzymatic activity sufficient to reduce PKU symptoms. More than 50% of PAH mutations described so far are missense, and the most frequent in severe PKU is the c.1222C>T (p.Arg408Trp) single nucleotide variant, with the p. [Arg408Trp]; [Arg408Trp] as the most common genotype (14). In 2016, Pan and colleagues attempted the first gene editing approach aimed to correct the c.1222C>T variant: in particular, they employed a modified CRISPR system with a deactivated Cas9 (dCas9) and an RNA-guided FokI nuclease as a tool in order to repair this genetic defect in an in vitro PKU model established by themselves, a c.1222C>T Cos7 cell line. The authors demonstrated that about 27% of analyzed cell clones exhibited the correction of the variant, and no evidences of off-target cleavages. Moreover, PAH activity was also partially rescued (15). More recently, gene editing of another PAH pathogenic variant (c.835T>C; p.F263S) has been attempted, taking advantage of the existence of an animal model, the homozygous Pahenu2 mouse, harboring in homozygosis this missense mutation (16). By using CRISPR-associated-cytidine deaminase base editors with rAAV-based delivery, Villiger research group reached a mRNA correction rate up to 63%, that restored physiological phenylalanine blood level in adult mice. In addition, the growth retardation normally observed in Pahenu2 animals was reduced in corrected heterozygous mice with respect to their homozygous Pahenu2 littermates (17). This result suggests the applicability of base editors for gene correction in the adult, particularly in hepatic tissue that is characterized by reduced proliferative capability.
OTC deficiency
Ornithine transcarbamylase (OTC) deficiency is an X-linked inborn error of metabolism of the urea cycle. The disease is due to pathogenic variants in the OTC gene that result in a decreased expression or reduced activity of the OTC enzyme. As for others urea cycle disorders, OTC deficiency is characterized by the presence of hyperammonemia, encephalopathy, and respiratory alkalosis. Clinically, the disease can occur with severe neonatal-onset (in males) that manifests with early and repeated, often lethal, metabolic crises, or as a post-natal onset disease, characterized by a partial enzyme deficiency, in both males and females. The long-term treatment of the disease includes restriction of protein intake and use of ammonia scavengers such as sodium benzoate. Liver transplantation is taken into consideration if hyperammonemic crises are life-threatening (18). OTC deficiency has been a target for a rAAV-mediated CRISPR/Cas9 gene correction: in newborn mice with a partial enzymatic deficiency [spfash mouse, carrying a G>A point mutation at donor splice site at the end of exon 4, resulting in aberrant mRNA splicing and reduced level of OTC transcript and protein (19)], this approach exerted HDR and reverted the mutation in about 10% of hepatocytes. This resulted in an increased survival in mice with high-protein diet that is usually a trigger of hyperammonemia. By converse, in adult mice the gene correction rate was remarkably lower, with the occurring of unexpected large deletions in DNA that resulted in toxicity and adverse effects in treated animals. These findings seem to indicate that non-dividing adult hepatocytes might have NHEJ mechanisms different from those of dividing newborn hepatic cells, thus affecting the DNA repair (20).
Arginase-1 deficiency
Arginase-1 deficiency, or argininemia, is another inborn error of metabolism involving a critical step in urea cycle, the hydrolysis of arginine to urea and ornithine. The inheritance of this disease, due to pathogenic variants in the ARG1 gene, is autosomal recessive. As for other urea cycle disorders, the clinical management of this disease is mainly based on severe restriction in protein dietary intake, that may be associated to treatment with nitrogen scavengers. Enzyme replacement therapies have been also attempted (21). Different mouse models of arginase-1 deficiency have been recently established (22-24). In particular, Sin and colleagues generated an inducible mouse model via Cre-mediated excision of exons 7 and 8 of Arg1 gene and subsequently attempted to correct the genetic defect by a CRISPR/Cas9 system associated to a piggyBac technology in induced pluripotent stem cells (iPSCs)-derived mouse hepatocytes and macrophages. Gene repair was successfully achieved in iPSCs, but unfortunately the differentiated hepatocytes did not show sufficient urea cycle function recovery, probably due to an inadequate cell maturation. By converse, iPSCs fully differentiated in macrophages exhibited substantial amounts of arginase-1 expression (25). The same research group performed a TALEN-mediated reincorporation of deleted exons in iPSCs from their murine model: successfully edited cells were differentiated in hepatocyte-like cells, and transplanted in the liver of the arginase-1 deficient mouse. Nevertheless, the arginase-1 deficiency phenotype was not adequately rescued, since there were non-optimal engraftment and insufficient hepatic repopulation (26). In summary, these studies constitute a proof-of-principle for gene correction in arginase-1 deficiency, and underline the need for further optimization of hepatocyte-like cells maturation and liver repopulation protocols.
Alpha-1 antitrypsin deficiency (AATD)
Alpha-1 antitrypsin deficiency is an autosomal recessive metabolism disorder, whose prevalence is particularly high in North America and in Scandinavia. The disease is due to biallelic pathogenic variants in SERPINA1, the gene encoding for α-1antitrypsin (AAT). The most prevalent SERPINA1 mutation is c.1096G>A, leading to the p.Glu342Lys substitution, the so-named Z protein variant. The most common clinical manifestation of AATD is chronic obstructive pulmonary disease (COPD). Affected individuals may also have hepatic dysfunction that typically presents with jaundice, hyperbilirubinemia and high serum aminotransferase level. Lung disease is due to a reduced inhibition of elastase, resulting in a higher elastin degradation in alveolar walls. By converse, the hepatic phenotype derives from an accumulation of mutant AAT that polymerizes in the hepatocytes. To date, the management of AATD-related lung disease relies on injections of plasma enriched for AAT, while hepatic disease often leads patients to the need of liver transplantation (27). The proof-of-principle of the feasibility of gene editing to correct AATD phenotype has been provided from Yusa group that employed a combination of ZFNs and piggyBac transposon technology to achieve homozygous correction for the c.1096G>A SERPINA1 point mutation in hiPSCs from an AATD patient (28,29). Another research group focused on AATD patient-derived hiPSCs using CRISPR/Cas9, demonstrating that Cas9 specifically targeted the wild-type or the mutant allele with only background levels of indels at the other allele, thus underlining the potential of CRISPR/Cas9 system for allele-specific genome editing (30). Indeed, CRISPR/Cas9 successfully corrected the Glu342Lys mutation in PiZ mouse, a model that recapitulates the human AATD hepatic phenotype since it expresses human Glu342Lys AAT (hAAT) as a transgene (31): in both neonatal and adult mice, gene editing partially rescued normal AAT serum level, even if the suggested therapeutic threshold was not reached (32). Similar results in PiZ mouse were obtained also by targeting with CRISPR/Cas9 the exon 2 of human SERPINA1, to disrupt the transcription of the transgene: AAT expression was reduced more than 98% in hepatocytes, leading to reduction of mutant AAT hepatic accumulation. In order to address also lung symptoms in AAT patients, a dual rAAV system enabling the correction of the Glu342Lys variant through HDR was also delivered, but a modest level of targeted gene correction (5%) was obtained (33). Bjursell and colleagues also employed the PiZ mouse and targeted the human mutant SERPINA1 gene in liver with CRISPR/Cas9: they achieved a disruption of human transgene that led to a reduction in plasma and liver hAAT level, hepatic protein aggregation and liver fibrosis (34).
In another approach, a promoterless AAV vector expressing wild-type AAT together with a synthetic miRNA targeting the pathogenic allele was integrated in the genome of hepatocytes used for transplantation in the modified NSG-PiZ mouse model, derived from PiZ and immune-deficient (NSG) mouse strains. The treatment improved the hepatic phenotype of mice, due to the increased level of normal AAT and to a concomitant decrease in mutant protein (35). Another application of CRISPR/Cas9 for correction of the AATD phenotype has been attempted in order to obtain targeted integration of human SERPINA1 gene in the Rosa26 safe harbor locus (i.e., a transgene insertion site that causes no apparent adverse effects on fitness, and permits stable gene expression) in C57BL/6J murine liver: this knock-in achieved a long-term augmentation of AAT serum level (36). Overall, these research studies highlight a raising interest in the development of preclinical models of genome editing therapies for AATD.
Tyrosinemia type 1 (HT1)
HT1 is a rare inborn error of tyrosine catabolism caused by defective activity of fumarylacetoacetate hydrolase (FAH), an enzyme that catalyzes the final step in the metabolism of fumarylacetoacetate. Mutations in the FAH gene, inherited as autosomal recessive, result in the deficiency of FAH, with consequent accumulation of products such as fumaryl- and maleyl-acetoacetate and their derivatives, leading to severe liver damage and renal tubular dysfunction. Nitisinone, the only pharmacological option available, acts by inhibiting, in the metabolic pathway at the second step of tyrosine catabolism upstream of FAH, the hydroxyphenylpyruvate dioxigenase (HPD), rescuing the phenotype and preventing acute liver injury (37). HT1 is a particularly suitable model for gene repair-based therapy because the repaired hepatocytes have a selective advantage and can expand and repopulate the liver (38-40). The strategies applied so far for genome editing of HT1 had two main targets, the correction of FAH gene mutations and the knockout of HPD gene, in the so-called metabolic reprogramming. For correction of fah gene mutations without the use of nucleases, attempt to perform HDR in the fah gene by using rAAV containing genomic fah sequence was performed in a mouse model of HT1 (fah-/- mice), demonstrating that corrected hepatocytes can survive and repopulate the liver, and showing liver function improvements (41). A similar strategy achieved successful HR-mediated genome editing of a Fah gene expression cassette at the Rosa26 locus, showing robust repopulation of the liver as indicated by survival, weight gain and multiple large Fah+ cell clusters (42). In the same mouse model, in 2014 Yin and colleagues attempted to correct the disease-causing mutation by CRISPR/Cas9-exerted HDR. Hydrodynamic tail vein injection of a vector, co-expressing the sgRNA and Cas9, and the single-stranded DNA (ssDNA) homology sequence was performed in fah-/- mice, showing a functional rescue of the Fah deficiency-induced liver damage, demonstrated by a decrease of serum markers such as aspartate aminotransferase (AST), alanine aminotransferase (ALT) and bilirubin. Moreover, widespread patches of Fah+ hepatocytes (33.5%) were present in treated mice with an initial repair frequency of 0.40% (43). The same group obtained similar results but with an increased number of initial repair frequency (6%) by systemic delivery of Cas9 mRNA in LNPs and sgRNA/HDR template by rAAV2/8 (44). A similar strategy was also applied to a Fah mutant rat model. A two-AdV system was generated to deliver sgRNA/donor template and Cas9 nickase (Cas9n) into adult HT1 rats. The initial correction rate was very low (0.1%), but corrected hepatocytes expanded and reached 95% of the liver tissue after 9 months of treatment. The collagen deposition was significantly narrowed compared with rats treated for 3 months, suggesting that liver fibrosis, the key chronic manifestation of human HT1, not detectable in the mouse model, had already ceased (45). An alternative strategy to HDR is represented by microhomology-mediated end-joining (MMEJ) for which a microhomologous sequence (5–25 bp) triggers error-prone end-joining (46). Cas9 together with Fah-MMEJ constructs were injected in a fah−/− mouse model, with the aim to insert Fah cDNA, exon 5–14, into intron 4 of Fah gene. Treated Fah−/− mice showed reduced body weight loss and a significantly reduced liver damage as indicated by decreased AST and ALT serum levels (47). In an alternative MMEJ-based approach, a targeted sequence substitution method was performed involving the use of two gRNAs, targeting the regions at the front and back of exon 8 of Fah gene, and the donor plasmid composed by the wild-type exon 8 flanked by 40-bp microhomology arms. Substitution rate in Fah-/-mice was about 5% and showed widespread patches of FAH+ hepatocytes (64.42%) (48). A completely different approach was attempted in a compound heterozygous mouse model of HT1, the Fahneo/PM mice, and used Cas9 to create DNA DSBs in both chromosomal homologs, thereby inducing allelic exchange between two different mutant alleles and rescuing the disease phenotype. Fahneo/PM mice were systematically treated with rAAVs: one harboring the Cas9 and the other a sgRNA targeting Fah intron 7. Fah gene expression was restored in both newborn and young adult treated mice, suggesting that allelic exchange can occur in hepatocytes beyond the fast-proliferating, postnatal stage (49). Gene repair using CRISPR-Cas9 has demonstrated curative therapy in animal models of liver disease also with ex vivo corrected hepatocytes. In two similar approaches performed by the same group, either Lentiviral vectors or rAAV were applied to deliver the sgRNAs and the Streptococcus pyogenes Cas9 (spCas9) or Staphylococcus aureus Cas9 (saCas9), respectively. In both cases, a rAAV was used to deliver the homology template to Fah-/- cells in vitro. Corrected hepatocytes were transplanted into syngeneic Fah-/- mice by intrasplenic injection. Mice displayed significant repopulation with corrected hepatocytes, and tyrosine levels in the blood were reduced to normal levels although, as expected, the non-integrative approach showed less on target indels (50,51). The metabolic pathway reprogramming is the other genome-editing-based strategy applied for HT1 and relies on silencing of Hpd gene. In the first reported approach, a genetic deletion of a critical exon of Hpd in the liver was applied using CRISPR/Cas9. Editing efficiencies reached 92% after 8 weeks of treatment, and Fah-/-/Hpd-/- hepatocytes demonstrated to have a growth advantage over non-edited hepatocytes, replacing the entire liver in only a few weeks. Treated animals gained weight and showed decreased levels of tyrosine in the plasma, confirming the potential therapeutic effect of this strategy (52). Similar results were obtained by delivering Neisseria meningitides Cas9 (NmeCas9) and its sgRNA by an all-in-one rAAV (53) or by Nanoblades, a delivery system of Cas9 ribonucleoproteins based on murine leukemia virus (54). Finally, in a recent approach, Rossidis and coworkers applied a BE technology to introduce in utero a nonsense mutation in the Hpd gene to permanently knock-out gene function. Base editing in the liver reached 40% with low rates of on-target indels. The in utero treatment rescued the lethal phenotype in Fah–/– mice and demonstrated a significantly improved liver function (55).
Mucopolysaccharidosis
Mucopolysaccharidoses are a group of diseases belonging to the family of lysosomal storage disorders. Gene editing approaches described so far target mucopolysaccharidosis type 1 (MPS1) and type 2 (MPS2). MPS1 is a progressive multisystem disorder with autosomal recessive inheritance, and it is caused by mutations of the IDUA gene, encoding a glycosidase involved in lysosomal degradation of glycosaminoglycans (GAG). Subjects with severe MPS1 have as main clinical features progressive skeletal dysplasia, severe intellectual disability and hearing loss. Death due to cardiorespiratory failure usually occurs in childhood (56). MPS2 is an X-linked recessive disease, due to absent or reduced levels of another enzyme involved in GAG catabolism, encoded by the IDS gene. The disease shows wide variability in age of onset, rate of progression and severity (e.g., of CNS involvement, airway and cardiac disease, skeletal abnormalities) (57). A platform for genome editing of hepatocytes has been tested for both MPS1 and 2 and involved ZFN-mediated site-specific integration of the cDNA of their corresponding defective genes within intron 1 of the safe harbor Alb gene (Alb). This strategy allowed in wild- type mice to recover expression of lacking enzymes (58), and proved its effectiveness and potential as a therapeutic strategy for treatment of the MPS in MPS2 mice (IDSy/-) (59). In this model, rAAV2/8 encoding a pair of ZFNs were targeted at the Alb locus along with a hIDS donor. Both IDS plasma levels and enzymatic activity increased with time and in a dose-dependent manner. The protein levels persisted through time and were found in other organs thus demonstrating that stable elevated levels of circulating IDS could be achieved with a single treatment, and that IDS is released from the liver into the circulation, from which it is up-taken by other secondary organs and tissues with subsequent reduction of GAG storage (59). Two clinical trials are already investigating this therapeutic approach for both MPS1 and 2 in the first ever evaluation of in vivo genome editing in humans (NCT03041324 and NCT02702115). Moreover, as CRISPR/Cas9 is approaching the clinical practice, in vitro studies on MP1 patient’s fibroblasts successfully demonstrated a CRISPR/Cas9-mediated HDR targeted at IDUA gene. This system was delivered by transfection with different liposomes and led to an increase of IDUA expression in corrected cells (60-62). Moreover, iPSCs generated from an Idua knockout mouse were targeted with CRISPR/Cas9 and gene-corrected iPSC-derived fibroblasts demonstrated enzyme function equivalent to the wild-type iPSC-derived fibroblasts (63). When applied to a newborn MPS1 mice model, a CRISPR/Cas9 strategy aimed at the insertion of IDUA gene in the Rosa26 locus led to increased IDUA levels in different organs and to reduced GAG accumulation with improvement in cardiovascular parameters (61). Alternative approaches such as the already mentioned allelic exchange proved to be successful in a mouse model of MPS1 heterozygous compound (49).
Hemophilia
Hemophilia is an X-linked congenital bleeding disorder characterized by severe bleeding episodes. The disease is caused by mutations in the F8 gene encoding coagulation factor VIII (FVIII), or in the F9 gene encoding coagulation factor IX (FIX), which are the causes of hemophilia A (HA) and B (HB), respectively. HA, one of the most common genetic bleeding disorders, is caused by various genetic mutations, which include large deletions, insertions, inversions, and point mutations in the F8 gene. Approximately 50% of severe HA cases are caused by two different types of chromosomal inversions that result from non-allelic HDR involving sequences present in intron 1 or 22 and their corresponding homologous sequences located far upstream of the F8 gene (64-66). First attempts to correct genetic defects through genome editing were aimed at targeting these types of inversions. Either CRISPR/Cas9 or TALENS were applied to perform the flip-flop of both inversions in hiPSCs harboring the genetic defects (67,68). Reversion efficiency was up to 6.7% and allowed expression of wild-type F8 in hiPSCs-derived endothelial cells. Moreover, when these endothelial cells were transplanted into HA mice, F8 enzymatic activity was significantly higher than that in non-transplanted mice (68). A different strategy to correct the inversion used TALENs to insert, via HDR, the exon 23–26 fragment of F8 cDNA, precisely at the junction of exon 22-intron 22 in HA-hiPSCs. Both transcript and protein expression were rescued, as demonstrated in corrected hiPSCs-derived endothelial cells (69). Universal approaches to correct all genetic variants found in HA patients were reported by the same and other groups. These strategies are characterized by nuclease-mediated gene addition of F8 cDNA fragment into FVIII locus, H11 safe harbor or Alb locus (58,70,71). In the first demonstration that FVIII locus is a suitable site for integration of the normal F8 gene, Sung and colleagues introduced into HA-hiPSCs a B-domain deleted form of F8 gene under the EF1α promoter via CRISPR/Cas9, demonstrating a significant increase in FVIII activity in knocked-in hiPSCs-derived endothelial cells (70). Similar results were obtained by the same group when FVIII gene was knocked-in via CRISPR/Cas9 into the human H11 safe harbor of both deleted and inverted HA-hiPSCs (71), further demonstrating that this strategy may provide a therapeutic approach for HA patients. Similarly to what described for MPS1 and 2, an in vivo validation of the feasibility of these universal approaches was the site-specific integration of F8 gene into Alb locus. In this approach, HA/CD4 null mice were treated with ZFNs pairs, individually packaged into rAAV8, targeted at intron 1 of the Alb gene, and a donor rAAV encoding a truncated form of hFVIII. This strategy resulted in a significantly increased hFVIII activity (37%) able to correct the activated partial thromboplastin time (aPTT) (58). The same approach was also applied to HB, an ideal disease for a liver-directed genome editing strategy since modest levels of hFIX activity can greatly improve the disease phenotype. Here the cDNA of F9 exon 2–8 flanked by a splicing acceptor signal and a poly-A were delivered together with rAAV8-ZFNs to mice via tail vein injection. Although the hybrid mAlb-hF9 mRNA represented a small fraction of total wild-type mAlb transcript, substantial levels of hFIX were obtained and were stable for up to 1 year, thus demonstrating that few hepatocytes need to be corrected to obtain high levels of FIX in the blood and that a single treatment can exert long term effects. Indeed, by treating HB mice with the same system, FIX enzymatic activity restoration was confirmed, as long as the recovery of the correct aPTT (58). The same strategy was applied by delivering ZFNs mRNAs by LNPs, not limited by the presence of preexisting neutralizing antibodies (72). In December 17, 2018 Sangamo Therapeutics announced the treatment of the first patient (Clinical Trial: NCT02695160) using SB-FIX, a therapeutic for ZFN-mediated genome editing delivered by rAAV intended to insert the corrective copy of the Factor IX cDNA into the Alb locus. Attempts to exploit the same mAlb locus via rAAV-exerted HR without the use of nucleases were performed by Barzel and colleagues. In this strategy, the authors knocked-in full F9 cDNA preceded by a 2A sequence upstream of the stop codon of mAlb in neonatal and adult mice showing an increased FIX plasma level and a normal coagulation time when F9 deficient mice were treated (73). Alternative universal knock-in strategies applied to HB mice models involved the transgene insertion in F9, AAVS1 or Rosa26 loci (74-77). In the first approach, an insertion of F9 cDNA encoding exons 2–8 was introduced via rAAV8-packaged-ZFNs into intron 1 of F9 gene, thus leading to HR and FIX increased levels with significantly shortened aPTT. In both neonatal and adult hF9/HB mice this approach was very effective, although with a 5 fold higher expression of FIX in adult animals probably imputable to a lower loss of rAAV (74,75). In two similar strategies, CRISPR/Cas9 were applied to HB-derived hiPSCs and a juvenile model of HB to insert full F9 sequence in, respectively, AAVS1 and Rosa26 loci. An increased expression of F9 transcript and hFIX activity was observed in F9-AAVS1-hiPSCs-derived hepatocytes (76) as well as in F9-Rosa26-juvenile R333Q hemophilia mice (77). Finally, three very similar approaches attempted to correct F9 gene in a mutation-specific manner by using HR exerted by CRISPR/Cas9, either via naked plasmids, AdV and rAAV8, in HB mice models (78-80). All three attempts reached an increase in transcripts and protein F9 expression with shortened aPTT, demonstrating the feasibility of this strategy even though AdV showed a higher hepatic injury (78). Moreover, Cas9 proteins demonstrated to be more suitable for germline gene therapy, respect to the use of mRNAs, leading to higher gene recovery rates, less embryo toxicity, and lower mosaic repair percentage (79).
Hypercholesterolemia
Hypercholesterolemia is a condition characterized by very high levels of cholesterol in the blood that lead to increased risk of developing coronary heart disease (CHD). Even if statin therapy is very effective, there is still large residual risk of CHD, and many patients are intolerant to statin therapy. Proprotein convertase subtilisin/kexin type 9 (PCSK9) has recently emerged as a promising therapeutic target for the prevention of CHD. PCSK9 functions as an antagonist to the LDL receptor, and while gain-of-function mutations lead to high levels of LDL-Cholesterol (LDL-C) and premature CHD (81), individuals with single loss-of-function mutations have a significant reduction of CHD risk (82,83). Moreover, individuals with two loss-of-function mutations do not suffer adverse clinical effects (84,85). All these observations suggested that therapies directed against PCSK9 would offer cardiovascular benefit, and PCSK9-targeting monoclonal antibodies are already in clinical trials (86). Meanwhile, many efforts are ongoing to apply new technologies such as genome editing to allow silencing of PCSK9 gene expression by targeted NHEJ, base editing and transcriptional silencing. The first attempt to support the knockout strategy of PSCK9 by NHEJ was performed by Ding and colleagues which designed a spCas9-based system targeting exon1 of murine pcsk9 and delivered it to wild-type mice by adenoviral vector (AdV) transduction obtaining up to 50% of editing efficiency with significant reduction of plasma PCSK9 and cholesterol (87). A very similar approach with comparable results was adopted by the same group by transplantation of primary human hepatocytes in Fah-/- chimeric humanized mice with the target of the sgRNA being human PCSK9 (hPCSK9) (88). In order to further enhance the efficiency of the system, Yin and colleagues combined the spCas9 approach (in form of mRNA), with a chemically modified sgRNAs, termed enhanced sgRNA, to be delivered intravenously in mice by LNPs. Through this very efficient system, this group was able to reach 83% of editing efficiency, undetectable level of plasma mpcsk9 and reduction of cholesterol levels up to 40% with no detectable off-target events and liver toxicity (89). This lipid-based delivery system has been applied also for the delivery of a PCSK9-targeted ZFNs-mRNAs, resulting in 90% reduction of protein plasma levels (72). In an attempt to exploit rAAV for delivery of PCSK9-targeted Cas9 system, three different groups used compact Cas9 orthologs that can be packaged in all-in-one rAAV: SaCas9, and two different type of NmeCas9 and Nme2Cas9 (53,90,91). All three groups delivered these systems into mice and obtained a very similar range of indels frequency (35–40%), very similar downregulation of Pcsk9 in serum and a decrease of about 40% in cholesterol. No sign of liver damages or inflammation were reported, suggesting that Cas9 orthologs may represent an alternative strategy to spCas9 to allow delivery by liver-tropic rAAV. rAAV8 have also been chosen to inactivate PCSK9 by meganuclease-mediated DSBs in macaques, a more relevant animal model. In this study, the knockout of PCSK9 resulted in a stable reduction in circulating PCSK9 and serum cholesterol and, by using a second-generation PCSK9-specific meganuclease, reduced off-target cleavage, highlighting safety considerations for clinical translation (92). An alternative knock-out strategy has been recently proposed based on BE. This approach has been tested on mPcsk9 by identifying a particular codon for W159, with possible alteration resulting in W159X alleles. A single AdV inoculation was used to deliver the BE and the sgRNA, and a substantial base editing activity in the liver was observed with a reduction in plasma PCSK9 and cholesterol levels of 28% and 56%, respectively. On average, 22% of alleles were specifically edited to W159X whereas indels rate was about 1% and no off-target effects were found (93). This strategy was applied also for in utero editing and in a humanized mouse model of hPCSK9 (55,94). In the first case, the on-target efficiency was about 10–15%, with less than 2% of indels. Whereas no off-target effect was observed, postnatal levels of PCSK9 were significantly reduced in parallel with cholesterol, and these effects were stable over time (55). Since mouse W159 is also proximal to a loss-of-function variant of hPCSK9, the same strategy was applied to the hPCSK9 knock-in mouse showing comparable results in terms of hPCSK9 and cholesterol downregulation with an on-site editing efficiency of 10–31% (94). An alternative approach to silence Pcsk9 has been proved by the use of a dCas9 fused to the transcriptional modulator Krüppel-associated box epigenetic repressor motif (KRAB, dCas9KRAB). A dual rAAV8 system harboring the dSaCas9KRAB repressor and the Pcsk9-sgRNA was injected into mice and showed a significant transcriptional silencing with no direct off-target effect on gene expression. Levels of Pcsk9 and cholesterol were decreased and the effects were sustained through time (95) thus demonstrating the feasibility and efficacy of this approach.
Conclusions
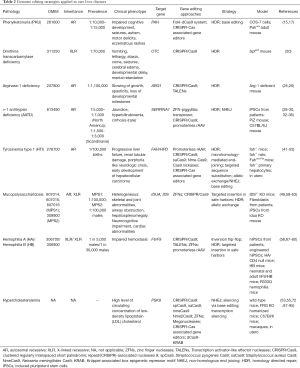
With the first in vivo ZFNs-based clinical trials for HB and MPS already on the run, genome editing is facing a new pragmatic era that only with time will reveal more about the safety and efficacy of these therapeutic approaches in humans. The liver has been targeted with traditional gene therapy for a while now, proving that it is an ideal organ for these types of therapeutics but also that the different outcomes depend on its ability to regenerate, hence diluting episomal transgenes by cell division, especially in younger patients. More definitive and sustained efficiencies are expected by engineered nuclease-based approaches (summarized in Table 2 of this review) and this theory is corroborated by many several aspects: (I) most rare liver diseases are caused by the lack of functioning proteins, with the restoration of even low levels being able to suffice for disease correction; (II) in some type of liver diseases, i.e., tyrosinemia type I, corrected hepatocytes have a growth advantage; (III) targeted integration of transgenes in safe harbors such as Alb locus have showed high level of expression and secretion of desired proteins. Moreover, with the very recent introduction of base editors, a further step toward single nucleotide-precise correction has been made. Nevertheless, such technology is still in its infancy and many efforts need to be made in order to reduce off-target effects and to refine the system to reach the clinical phase.
Full table
Acknowledgments
We would like to acknowledge Greta Bernardo for her skillful technical assistance.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Hackl C, Schlitt HJ, Melter M, et al. Current developments in pediatric liver transplantation. World J Hepatol 2015;7:1509-20. [Crossref] [PubMed]
- Colella P, Ronzitti G, Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol Ther Methods Clin Dev 2017;8:87-104. [Crossref] [PubMed]
- Finn JD, Smith AR, Patel MC, et al. Report A Single Administration of CRISPR / Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing Report A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep 2018;22:2227-35. [Crossref] [PubMed]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013;31:397-405. [Crossref] [PubMed]
- Stoddard BL. Homing endonucleases: From microbial genetic invaders to reagents for targeted DNA modification. Structure 2011;19:7-15. [Crossref] [PubMed]
- Trevisan M, Palù G, Barzon L. Genome editing technologies to fight infectious diseases. Expert Rev Anti Infect Ther 2017;15:1001-13. [Crossref] [PubMed]
- Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016;533:420-4. [Crossref] [PubMed]
- Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 2018;19:770-88. [Crossref] [PubMed]
- Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017;551:464-71. [Crossref] [PubMed]
- Jin S, Zong Y, Gao Q, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 2019;364:eaaw7166.
- Zuo E, Sun Y, Wei W, et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 2019;364:eaav9973.
- Blau N, Spronsen FJ, Van , Levy HL. Phenylketonuria. Lancet 2010;376:1417-27. [Crossref] [PubMed]
- Strisciuglio P, Concolino D. New strategies for the treatment of phenylketonuria (PKU). Metabolites 2014;4:1007-17. [Crossref] [PubMed]
- Blau N, Shen N, Carducci C. Molecular genetics and diagnosis of phenylketonuria: state of the art. Expert Rev Mol Diagn 2014;14:655-71. [Crossref] [PubMed]
- Pan Y, Shen N, Jung-Klawitter S, et al. CRISPR RNA-guided FokI nucleases repair a PAH variant in a phenylketonuria model. Sci Rep 2016;6:35794. [Crossref] [PubMed]
- Shedlovsky A, McDonald JD, Symula D, et al. Mouse models of human phenylketonuria. Genetics 1993;134:1205-10. [PubMed]
- Villiger L, Grisch-Chan HM, Lindsay H, et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med 2018;24:1519-25. [Crossref] [PubMed]
- Lichter-Konecki U, Caldovic L, Morizono H, et al. Ornithine Transcarbamylase Deficiency. GeneReviews 1993-2019; University of Washington, Seattle.
- Hodges PE, Rosenberg LE. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc Natl Acad Sci 1989;86:4142-6. [Crossref] [PubMed]
- Yang Y, Wang L, Bell P, et al. Metabolic Liver Disease in Newborn Mice. Nat Biotechnol 2016;34:334-8. [Crossref] [PubMed]
- Diez-Fernandez C, Rüfenacht V, Gemperle C, et al. Mutations and common variants in the human arginase 1 (ARG1) gene: Impact on patients, diagnostics, and protein structure considerations. Hum Mutat 2018;39:1029-50. [Crossref] [PubMed]
- Sin YY, Ballantyne LL, Mukherjee K, et al. Inducible Arginase 1 Deficiency in Mice Leads to Hyperargininemia and Altered Amino Acid Metabolism. PLoS One 2013;8:e80001. [Crossref] [PubMed]
- Kasten J, Hu C, Bhargava R, et al. Lethal phenotype in conditional late-onset arginase 1 deficiency in the mouse. Mol Genet Metab 2013;110:222-30. [Crossref] [PubMed]
- Burrage LC, Sun Q, Elsea SH, et al. Human recombinant arginase enzyme reduces plasma arginine in mouse models of arginase deficiency. Hum Mol Genet 2015;24:6417-27. [Crossref] [PubMed]
- Sin YY, Price PR, Ballantyne LL, et al. Proof-of-Concept Gene Editing for the Murine Model of Inducible Arginase-1 Deficiency. Sci Rep 2017;7:2585. [Crossref] [PubMed]
- Sin YY, Ballantyne LL, Richmond CR, et al. Transplantation of Gene-Edited Hepatocyte-like Cells Modestly Improves Survival of Arginase-1-Deficient Mice. Mol Ther Nucleic Acids 2018;10:122-30.
- Stoller JK, Lacbawan FL, Aboussouan LS. Alpha-1 Antitrypsin Deficiency. GeneReviews 1993-2019; University of Washington, Seattle.
- Yusa K, Rashid ST, Strick-marchand H, et al. Targeted gene correction of α 1 -antitrypsin deficiency in induced pluripotent stem cells. Nature 2011;478:391-4. [Crossref] [PubMed]
- Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc 2013;8:2061-78. [Crossref] [PubMed]
- Smith C, Abalde-Atristain L, He C, et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol Ther 2015;23:570-7. [Crossref] [PubMed]
- Carlson JA, Barton Rogers B, Sifers RN, et al. Accumulation of PiZ α1-antitrypsin causes liver damage in transgenic mice. J Clin Invest 1989;83:1183-90. [Crossref] [PubMed]
- Song CQ, Wang D, Jiang T, et al. In Vivo Genome Editing Partially Restores Alpha1-Antitrypsin in a Murine Model of AAT Deficiency . Hum Gene Ther 2018;29:853-60. [Crossref] [PubMed]
- Shen S, Sanchez ME, Blomenkamp K, et al. Amelioration of Alpha-1 Antitrypsin Deficiency Diseases with Genome Editing in Transgenic Mice. Hum Gene Ther 2018;29:861-73. [Crossref] [PubMed]
- Bjursell M, Porritt MJ, Ericson E, et al. Therapeutic Genome Editing With CRISPR/Cas9 in a Humanized Mouse Model Ameliorates α1-antitrypsin Deficiency Phenotype. EBioMedicine 2018;29:104-11. [Crossref] [PubMed]
- Borel F, Tang Q, Gernoux G, et al. Survival Advantage of Both Human Hepatocyte Xenografts and Genome-Edited Hepatocytes for Treatment of α-1 Antitrypsin Deficiency. Mol Ther 2017;25:2477-89.
- Stephens CJ, Kashentseva E, Everett W, et al. Targeted in vivo knock-in of human alpha-1-antitrypsin cDNA using adenoviral delivery of CRISPR/Cas9. Gene Ther 2018;25:139-56. [Crossref] [PubMed]
- Lindstedt S. Treatment of hereditary tyrosinaemia type I by inhibition of 4-hydroxyphenylpyruvate dioxygenase. Lancet 1992;340:813-7. [Crossref] [PubMed]
- Aponte JL, Sega GA, Hauser LJ, et al. Point mutations in the murine fumarylacetoacetate hydrolase gene: Animal models for the human genetic disorder hereditary tyrosinemia type 1. Proc Natl Acad Sci 2001;98:641-5. [Crossref] [PubMed]
- Azuma H, Paulk N, Ranade A, et al. Robust expansion of human hepatocytes in Fah-/-/Rag2 -/-/Il2rg-/- mice. Nat Biotechnol 2007;25:903-10. [Crossref] [PubMed]
- Overturf K, Al-Dhalimy M, Tanguay R, et al. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet 1996;12:266-73. [Crossref] [PubMed]
- Paulk NK, Wursthorn K, Wang Z, et al. Adeno-associated virus gene repair corrects a mouse model of hereditary tyrosinemia in vivo. Hepatology 2010;51:1200-8. [Crossref] [PubMed]
- Junge N, Yuan Q, Vu TH, et al. Homologous recombination mediates stable Fah gene integration and phenotypic correction in tyrosinaemia mouse-model. World J Hepatol 2018;10:277-86. [Crossref] [PubMed]
- Yin H, Xue W, Chen S, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551-3. [Crossref] [PubMed]
- Yin H, Song C, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 2016;34:328-33. [Crossref] [PubMed]
- Shao Y, Wang L, Guo N, et al. Cas9-nickase–mediated genome editing corrects hereditary tyrosinemia in rats. J Biol Chem 2018;293:6883-92. [Crossref] [PubMed]
- Nakade S, Tsubota T, Sakane Y, et al. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat Commun 2014;5:5560. [Crossref] [PubMed]
- Yao X, Wang X, Liu J, et al. CRISPR/Cas9 – Mediated Precise Targeted Integration In Vivo Using a Double Cut Donor with Short Homology Arms. EBioMedicine 2017;20:19-26. [Crossref] [PubMed]
- Shin JH, Jung S, Ramakrishna S, et al. In vivo gene correction with targeted sequence substitution through microhomology-mediated end joining. Biochem Biophys Res Commun 2018;502:116-22. [Crossref] [PubMed]
- Wang D, Li J, Song CQ, et al. Cas9-mediated allelic exchange repairs compound heterozygous recessive mutations in mice. Nat Biotechnol 2018;36:839-42. [Crossref] [PubMed]
- VanLith C, Guthman R, Nicolas CT, et al. Curative Ex Vivo Hepatocyte-Directed Gene Editing in a Mouse Model of Hereditary Tyrosinemia Type 1 . Hum Gene Ther 2018;29:1315-26. [Crossref] [PubMed]
- VanLith CJ, Guthman RM, Nicolas CT, et al. Ex Vivo Hepatocyte Reprograming Promotes Homology-Directed DNA Repair to Correct Metabolic Disease in Mice After Transplantation. Hepatol Commun 2019;3:558-73. [Crossref] [PubMed]
- Pankowicz FP, Barzi M, Kim KH, et al. Rapid Disruption of Genes Specifically in Livers of Mice Using Multiplex CRISPR/Cas9 Editing. Gastroenterology 2018;155:1967-1970.e6. [Crossref] [PubMed]
- Ibraheim R, Song CQ, Mir A, et al. All-in-one adeno-associated virus delivery and genome editing by Neisseria meningitidis Cas9 in vivo. Genome Biol 2018;19:137. [Crossref] [PubMed]
- Mangeot PE, Risson V, Fusil F, et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat Commun 2019;10:45. [Crossref] [PubMed]
- Rossidis AC, Stratigis JD, Chadwick AC, et al. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat Med 2018;24:1513-8. [Crossref] [PubMed]
- Clarke LA. Mucopolysaccharidosis Type I. GeneReviews 1993-2019; University of Washington, Seattle.
- Scarpa M. Mucopolysaccharidosis Type II. GeneReviews 1993-2019; University of Washington, Seattle.
- Sharma R, Anguela XM, Doyon Y, et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood 2015;126:1777-84. [Crossref] [PubMed]
- Laoharawee K, DeKelver RC, Podetz-Pedersen KM, et al. Dose-Dependent Prevention of Metabolic and Neurologic Disease in Murine MPS II by ZFN-Mediated In Vivo Genome Editing. Mol Ther 2018;26:1127-36. [Crossref] [PubMed]
- Schuh RS, de Carvalho TG, Giugliani R, et al. Gene editing of MPS I human fibroblasts by co-delivery of a CRISPR/Cas9 plasmid and a donor oligonucleotide using nanoemulsions as nonviral carriers. Eur J Pharm Biopharm 2018;122:158-66. [Crossref] [PubMed]
- Schuh RS, Poletto É, Pasqualim G, et al. In vivo genome editing of mucopolysaccharidosis I mice using the CRISPR/Cas9 system. J Control Release 2018;288:23-33. [Crossref] [PubMed]
- de Carvalho TG, Schuh R, Pasqualim G, et al. CRISPR-Cas9-mediated gene editing in human MPS I fibroblasts. Gene 2018;678:33-7. [Crossref] [PubMed]
- Miki T, Vazquez L, Yanuaria L, et al. Induced pluripotent stem cell derivation and ex vivo gene correction using a mucopolysaccharidosis type 1 disease mouse model. Stem Cells Int 2019;2019:6978303. [Crossref] [PubMed]
- Lakich D, Kazazian HH, Antonarakis SE, et al. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 1993;5:236-41. [Crossref] [PubMed]
- Naylor J, Brinke A, Hassock S, et al. Characteristic mRNA abnormality found in half the patients with severe haemophilia A is due to large DNA inversions. Hum Mol Genet 1993;2:1773-8. [Crossref] [PubMed]
- Bagnall RD, Waseem N, Green PM, et al. Recurrent inversion breaking intron 1 of the factor VIII gene is a frequent cause of severe hemophilia A. Blood 2002;99:168-74. [Crossref] [PubMed]
- Park CY, Kim J, Kweon J, et al. Targeted inversion and reversion of the blood coagulation factor 8 gene in human iPS cells using TALENs. Proc Natl Acad Sci 2014;111:9253-8. [Crossref] [PubMed]
- Park CY, Kim DH, Son JS, et al. Functional Correction of Large Factor VIII Gene Chromosomal Inversions in Hemophilia A Patient-Derived iPSCs Using CRISPR-Cas9. Cell Stem Cell 2015;17:213-20. [Crossref] [PubMed]
- Wu Y, Hu Z, Li Z, et al. In situ genetic correction of F8 intron 22 inversion in hemophilia A patient-specific iPSCs. Sci Rep 2016;6:18865. [Crossref] [PubMed]
- Sung JJ, Park CY, Leem JW, et al. Restoration of FVIII expression by targeted gene insertion in the FVIII locus in hemophilia A patient-derived iPSCs. Exp Mol Med 2019;51:45. [Crossref] [PubMed]
- Park CY, Sung JJ, Cho SR, et al. Universal Correction of Blood Coagulation Factor VIII in Patient-Derived Induced Pluripotent Stem Cells Using CRISPR/Cas9. Stem Cell Reports 2019;12:1242-9. [Crossref] [PubMed]
- Conway A, Mendel M, Kim K, et al. Non-viral Delivery of Zinc Finger Nuclease mRNA Enables Highly Efficient In Vivo Genome Editing of Multiple Therapeutic Gene Targets. Mol Ther 2019;27:866-77. [Crossref] [PubMed]
- Barzel A, Paulk NK, Shi Y, et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature 2015;517:360-4. [Crossref] [PubMed]
- Li H, Haurigot V, Doyon Y, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011;475:217-21. [Crossref] [PubMed]
- Anguela XM, Sharma R, Doyon Y, et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood 2013;122:3283-7. [Crossref] [PubMed]
- Lyu C, Shen J, Wang R, et al. Targeted genome engineering in human induced pluripotent stem cells from patients with hemophilia B using the CRISPR-Cas9 system. Stem Cell Res Ther 2018;9:92. [Crossref] [PubMed]
- Stephens CJ, Lauron EJ, Kashentseva E, et al. Long-term correction of hemophilia B using adenoviral delivery of CRISPR/Cas9. J Control Release 2019;298:128-41. [Crossref] [PubMed]
- Guan Y, Ma Y, Li Q, et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med 2016;8:477-88. [Crossref] [PubMed]
- Huai C, Jia C, Sun R, et al. CRISPR/Cas9 - mediated somatic and germline gene correction to restore hemostasis in hemophilia B mice. Hum Genet 2017;136:875-83. [Crossref] [PubMed]
- Ohmori T, Nagao Y, Mizukami H, et al. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci Rep 2017;7:4159. [Crossref] [PubMed]
- Abifadel M, Benjannet S, Farnier M, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 2003;34:154-6. [Crossref] [PubMed]
- Cohen J, Pertsemlidis A, Kotowski IK, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 2005;37:161-5. [Crossref] [PubMed]
- Cohen JC, Boerwinkle E, Mosley TH, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 2006;354:1264-72. [Crossref] [PubMed]
- Zhao Z, Tuakli-Wosornu Y, Lagace TA, et al. Molecular Characterization of Loss-of-Function Mutations in PCSK9 and Identification of a Compound Heterozygote. Am J Hum Genet 2006;79:514-23. [Crossref] [PubMed]
- Hooper AJ, Marais AD, Tanyanyiwa DM, et al. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis 2007;193:445-8. [Crossref] [PubMed]
- Stein EA, Swergold GD. Potential of proprotein convertase subtilisin/kexin type 9 based therapeutics. Curr Atheroscler Rep 2013;15:310. [Crossref] [PubMed]
- Ding Q, Strong A, Patel KM, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res 2014;115:488-92. [Crossref] [PubMed]
- Wang X, Raghavan A, Chen T, et al. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo. Arterioscler Thromb Vasc Biol 2016;36:783-6. [Crossref] [PubMed]
- Yin H, Song CQ, Suresh S, et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol 2017;35:1179-87. [Crossref] [PubMed]
- Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015;520:186-91. [Crossref] [PubMed]
- Edraki A, Mir A, Ibraheim R, et al. A Compact, High-Accuracy Cas9 with a Dinucleotide PAM for In Vivo Genome Editing. Mol Cell 2019;73:714-726.e4. [Crossref] [PubMed]
- Wang L, Smith J, Breton C, et al. Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat Biotechnol 2018;36:717-25. [Crossref] [PubMed]
- Chadwick AC, Wang X, Musunuru K. In Vivo Base Editing of PCSK9 as a Therapeutic Alternative to Genome Editing. Arterioscler Thromb Vasc Biol 2017;37:1741-7. [Crossref] [PubMed]
- Carreras A, Pane LS, Nitsch R, et al. In vivo genome and base editing of a human PCSK9 knock-in hypercholesterolemic mouse model. BMC Biol 2019;17:4. [Crossref] [PubMed]
- Thakore PI, Kwon JB, Nelson CE, et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun 2018;9:1674. [Crossref] [PubMed]
Cite this article as: Trevisan M, Masi G, Palù G. Genome editing technologies to treat rare liver diseases. Transl Gastroenterol Hepatol 2020;5:23.


