
Gut microbiome changes in Nonalcoholic fatty liver disease & alcoholic liver disease
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease in developed countries. NAFLD is characterized by an excessive accumulation of lipids in the liver that is not caused by alcohol (1). NAFLD is closely associated with multiple risk factors such as metabolic syndrome, obesity, insulin resistance and dyslipidemia (1,2). Excessive caloric consumption above daily requirements, as well as energy-rich foods high in carbohydrates and fats have been shown to lead to the development of NAFLD (3). Interestingly, NAFLD is generally asymptomatic, and is usually diagnosed by a clinician after excluding common causes of elevated liver enzymes such as ALT and AST (4). While early stages of liver disease pose no serious health concerns and is reversible, NAFLD exists on a spectrum of liver diseases that could potentially progress to more serious pathologies including nonalcoholic steatohepatitis (NASH), liver fibrosis, cirrhosis and hepatocellular carcinoma (HCC) (5).
Alcoholic liver disease (ALD) is a common sequela from alcohol use disorder (AUD) and remains as one of the most common causes for a liver transplantation in the United States (6). The National Institute of Alcoholism and Alcohol Abuse (NIAAA) defines AUD as consumption of greater than 3 drinks per day in men and greater than 2 drinks per day in women (7). Similar to NAFLD, ALD exists as a spectrum of liver pathologies that range from hepatic steatosis to more advanced stages of disease including alcoholic hepatitis (AH), cirrhosis and HCC (8). Those that consume excessive amounts of alcohol that fit as AUD, up to 90% of these patients exhibit some degree of hepatic steatosis. However, only a fraction of these patients go on to develop AH and even a smaller percentage will advance to AH or cirrhosis (9).
NAFLD and ALD share many similar risk factors and disease pathologies. Various heritable genes, obesity and gender have been implicated in the severity of disease (10-14). In addition, endoplasmic reticulum stress, inflammation and the dysregulation of hepatic lipid metabolism has been shown to contribute to the pathophysiology of NAFLD and ALD (15,16). Both NAFLD and ALD have a similar disease spectrum of steatosis and steatohepatitis that can be associated with fibrosis, and can progress to more advanced stages of the disease including cirrhosis and liver cancer. Despite numerous research efforts in search of pharmacologic interventions, there are currently no FDA approved therapeutic agents for NAFLD and ALD. The mainstay treatment for patients is lifestyle modifications and abstinence from alcohol (17).
Recent studies have demonstrated the potential role of the gut microbiota in liver diseases (18,19). In the past decade, there has been a driving force to manipulate the gut microbiota and the composition of bile acids as potential drug targets as evidence suggests that the overgrowth of certain bacteria could lead to disease. This review will focus on the changes of the gut microbiome as it relates to NAFLD and ALD.
Gut microbiome
The human gut microbiome is a large reservoir of bacteria containing trillions of microorganisms in just a gram of intestinal content (20). Throughout evolution, humans have developed a symbiotic relationship with bacterial species that harbor the gut by providing the host essential vitamins and nutrients (21). Interestingly, the gut microbiome is vastly diverse and the composition of various genera and species are dependent on various factors such as environment, diet and host genetics (22,23). More importantly is that this delicate balance of commensal organisms residing in the host gastrointestinal tract can be disrupted which may lead to various diseases such as ALD, inflammatory bowel disease, Clostridium difficile infection, autism, Parkinson disease and various cancers including colorectal cancer (24-27). The shift towards an unfavorable population of microbiota is termed dysbiosis and can have profound effects on the overall health of the host. Given that the host environment and genetics play an important role in the microbiota make up and diversity, it is difficult to have one standard bacterial composition labeled as ideal and others as dysbiosis (28). However, studies have shown that certain commensal bacteria producing certain favorable compounds for the host such as short-chain fatty acids (SCFA) or anti-inflammatory substances are generally regarded as healthy whereas pro-inflammatory metabolites and endotoxins are harmful (29,30). The gut microbiota can have profound effects on the host immune system by eliciting the innate and adaptive immune responses through the production of various antigens, metabolites and toxins (31). In the context of liver disease, bile acids and endotoxins such as lipopolysaccharides (LPS) have been studied extensively and are implicated in disease pathogenesis (32).
With technological advances in genomics and bioinformatics, the human microbiome can be analyzed through next generation sequencing, 16S rRNA sequencing and a whole genome shotgun sequencing approach to sorting the diverse microbial communities (33,34). The organization of bacterial community diversity is based off of the hypervariable regions (V1-V9) of the bacterial 16S gene (35). This allows a consensus of the bacterial population from the host gut to group from phyla down to the genera of bacteria. Bacterial taxonomy can further be used to compare against existing databases and data mined to plot metagenomic scales of biodiversity and microbiome composition from various human bacterial reservoirs (36). Moreover, these data sets are organized into operational taxonomic unit (OTU) through mapping the 16S rRNA sequences which allows clustering of bacteria based on similar sequences (36). The organization based on OTU serves as a useful proxy for discriminating between species generally by sorting based on 99% similarity as the threshold (37). However, a drawback is that these techniques usually classify up to the bacterial genus level and that the set threshold often limits the ability to discriminate closely related species from different families such as Enterobacteriaceae, Clostridiaceae, and Peptostreptococcaceae (37).
Gut-liver axis and its role in liver disease
The gut and the liver interact through multiple avenues. Since blood drains from the gut back to the liver through the portal circulation, an intact gut barrier is critical in controlling bacteria, bacterial antigens and toxins such as LPS from reaching to the liver (38). The gut serves as both a physical and immunological barrier in containing harmful substances from the liver. The gut epithelium is held together by tight junctions and is coated by a mucosal layer of immunoglobulin (IgA) and is immunologically enhanced by mesenteric lymph nodes and Peyer’s patch to prevent significant translocation of bacterial antigens to the portal circulation (39). Alcohol has been shown to have profound effects on intestinal integrity and lead to a phenomenon known as “leaky gut” (40). Alcohol damages the intestinal epithelial lining allowing the translocation of bacteria and LPS into the portal circulation where resident liver macrophages known as Kupffer cells are sensitized and release pro-inflammatory cytokines such as IL-1β, IL-6, MCP-1 and TNFα (41). A recent study demonstrated that the disruption of SphK2 leads to a compromised intestinal epithelium and is exacerbated by alcohol in mice (42).
In addition, the gut interacts with the liver through the enterohepatic circulation whereby bile acids synthesized in the liver are secreted in the small intestine and reabsorbed in the ileum back to the liver (43). Humans synthesize mainly two primary bile acids (BAs)-cholic acid (CA) and chenodeoxycholic acid (CDCA). These primary BAs can further be conjugated by glycine or taurine to form deoxycholic acid (DCA) and lithocholic acid (LCA) (44). Bile acids serve as detergent molecules to aid in the digestion of fats and drug metabolism, and are potent signaling molecules (45). Studies have demonstrated that conjugated BAs regulate hepatic gene metabolism through the sphingosine 1-phosphate mediated signaling (46). In addition, the negative regulator of bile acid synthesis, farnesoid x receptor (FXR), has been shown to ameliorate liver injury, steatosis and inflammation from alcohol-induced liver injury (47).
Steatosis (fatty liver) is a common initial insult in individuals who abuse alcohol and individuals with obesity. The presence of hepatic steatosis can be associated with dysbiosis and increased intestinal permeability. In parallel, bile acid and choline homeostasis are disturbed along with increased translocation of microbial-associated molecular patterns (MAMPs) across the gut barrier, which can lead to steatohepatitis, the progressive form of liver damage. In the liver, MAMPs interact through pattern-recognition receptors (PRRs) on Kupffer cells and hepatic stellate cells, which perpetuate inflammation and fibrosis (1,2,48). Activation of Toll-like receptor (TLR) is the primary driver of immune response in liver disease, which are mediated by endotoxin for TLR4, methylated DNA for TLR9 and Gram-positive bacteria for TLR2 (1,2,48,49). Both ALD and NAFLD related liver disease can progress to cirrhosis. With progressive compromise in liver function, tumor- promoting metabolites and xenobiotics accumulate, which can activate oncogenic pathways associated with HCC, the most predominant form of primary liver cancers.
In summary, several common and overlapping key pathophysiologic processes link the microbiome to NAFLD and ALD. These include: (I) small intestinal bacterial overgrowth and altered microbiome composition; (II) altered gut barrier function and intestinal permeability; (III) close interaction of bile acids (BAs) with the host and microbiome, modulating microbiota, microbial metabolites and shifts in primary and secondary BAs profile; (IV) myriad of microbial associated metabolites (12,41,50,51).
Gut microbiota composition and changes in NAFLD
NAFLD is closely associated with obesity and diabetes (52). Insulin resistance has been shown to accelerate the accumulation of lipids in hepatocytes resulting in hepatic inflammation (53). The role of the gut microbiota in NAFLD is being actively studied, but causality has yet to be established. Among children and adolescents, compared to the healthy group, both obese and NASH groups show a significant increase in Bacteroides, mainly a result of >6-fold increase in family Prevotellaceae and genus Prevotella, as well as a marked decrease in Firmicutes, mostly driven by the decreased abundance in families Lachanospiraceae and Ruminococcaceae. A distinguishing feature of NASH from the obese group is enrichment in phylum Proteobacteria, family Enterobacteriaceae and genus Escherichia. This change is accompanied by a significantly elevated serum ethanol concentration in NASH patients alone (54). Moreover, NASH patients typically have bacterial overgrowth that may inhibit intestinal tight junction function and promote epithelial barrier dysfunction (12). In addition, bacterial overgrowth has been shown to upregulate hepatic toll-like receptor 4 (TLR4) and inflammatory cytokines such as IL-8 and TNF (10).
The microbiome composition in adults has some similarities and differences than children and adolescent. In children, relative abundance of Bacteroides is increased in adults, however the trend is opposite for adult NASH patients who show significantly increased Ruminococcus and markedly decreased Prevotella with stage 2 fibrosis or higher (55). Whole genome metagenomics in adult NAFLD patients reveal increased abundance of Escherichia coli, and Bacteroides vulgatus in patients with advanced fibrosis (stages 3 and 4) (56). These data indicate an association between gram-negative bacteria and progression of liver fibrosis. Also, a marked increase in genus Blautia and Megasphaera and a notable decrease in genus Bacillus is found in patients with NAFLD related cirrhosis (57).
Our knowledge regarding NAFLD and the gut microbiome, both at the observational and mechanistic levels, has grown. Leveraging this, microbiome is emerging as an attractive source of biomarkers for the diagnosis of NAFLD and related advanced fibrosis. As noted earlier, significantly elevated gut microbial production of ethanol distinguished obese children from NASH patients (54). NAFLD patients also show increased systemic TMAO and hepatic bile-acid synthesis, and decreased production of phosphatidylcholine (3,58-60). Recent works from Loomba et al. demonstrated that there are differences in carbon and amino acid metabolism in gut microbiome of patients with NAFLD-associated advanced fibrosis, and gut-microbiome-derived signature can detect NAFLD-cirrhosis (4,56). In the later study, a panel of 30 features, including 27 bacterial features derived using a Random Forest classifier model, can detect NAFLD-cirrhosis with a robust diagnostic accuracy (AUROC of 0.92), with similar results in a validation cohort of NAFLD-cirrhosis (AUROC of 0.87) (57). These data provide evidence for a fecal-microbiome derived signature to detect NAFLD fibrosis and cirrhosis as well as candidacy for potential anti-fibrotic treatment trials in NAFLD.
A diet consisting of choline has been shown to be associated with NAFLD through alterations in the gut microbiota composition that favor bacteria that breakdown choline (61). Choline is metabolized into dimethylamine and trimethylamine by gut bacteria and these compounds are converted into trimethylamine oxide in the liver that promotes liver inflammation and injury (62). C129S6 mouse studies demonstrated that a high-fat diet shifted the metabolome of the gut towards choline degradation (63). Moreover, an elegant mouse study involved the transplantation of gut microbiota from C57BL/6J mice that exhibited high fat diet (HFD)-induced NAFLD to germ-free (GF) mice (64). Subsequently, the GF mice developed NAFLD and signs of insulin resistance. Alterations in the gut microbiota have also been shown to play a role in the development of NASH. These studies underscore the importance of microbiome alterations in the development of NAFLD and other metabolic diseases.
Gut microbiota composition and changes in ALD
A number of experimental and human studies, have enriched our understanding of the compositional and mechanistic contributions of the gut microbiota in ALD. As in NAFLD, ALD may progress from simple steatosis to steatohepatitis (AH), and cirrhosis. During ALD progression, there is a shift in microbiota composition from AH to pre-cirrhosis to cirrhosis (19). Studies performed on chronic alcohol users found that bacterial overgrowth is key in disease progression towards AH and cirrhosis. Jejunal aspirates obtained from alcoholic patients showed increased numbers of aerobic and anaerobic bacteria (65). Colonic bacteria from alcoholic cirrhotic patients also showed a lower proportion of Bacteriodetes and higher proportions of Proteobacteria compared to alcoholic patients who did not develop cirrhosis (18). These findings are supported by mouse studies demonstrating that on a 3-week alcohol-fed diet, there are increased proportions of Bacteriodetes whereas Firmicutes were more prevalent in control mice (66). Interestingly, transfer of gut microbiota from patients who have AH to mice led to severe alcohol-induced liver inflammation injury (67). Interestingly, these changes correlated with endotoxemia. Also, alcohol-withdrawal or probiotic treatment can reverse alcohol-induced dysbiosis, but the response is only partial (68,69). These results highlight the potential role in the composition of the gut microbiota in promoting liver disease.
A recent study demonstrates a crucial role of gut microbiome in AH and that the disease severity of human AH can be transferred to germ-free and conventionalized mice (67). Importantly, fecal microbiota transplantation (FMT) from an alcoholic patient without AH improved the severe AH following the first FMT from the severe AH patient. An increase in certain species of Bifidobacteria, Streptococci and Enterobacteria along with a decrease in Clostridium leptum or Faecalibacterium prausnitzii, both well-established anti-inflammatory strains, in stool samples characterizes AH-related dysbiosis. These data suggest that human fecal AH samples contain proinflammatory signals which are transferable from humans to mice with the speculation that certain intestinal bacteria, i.e., pathobionts drive the overwhelming inflammatory processes of AH.
Bile acid composition in the gut has been linked to ALD progression. Chronic alcohol users have been shown to have alterations in bile acid synthesis and in the composition of the bile acid pool (50). In addition, bile acids could induce hepatic inflammation and promote liver fibrosis through the activation of hepatic stellate cells (70). FXR and TGR5 agonists demonstrated the potential to ameliorate liver steatosis, inflammation and injury in mice (47). Alcohol has been shown to induce bile acid synthesis in humans. The results are supported by patients with cirrhosis exhibited increased secondary bile acids compared to abstinent patients with cirrhosis (71). This is accompanied by increases in the pro-inflammatory mediators produced in the liver including IL-1β, IL-6, MCP-1 and TNFα.
Alcohol-associated dysbiosis in mice can reduce long chain fatty acids (LCFA) biosynthesis, and supplementation of LCFA can restore eubiosis. In fact, a significant correlation between Lactobacillus spp. and bacterial LCFA (C15:0 and C17:0) is seen in ALD patients but not in healthy controls (72). Also, alcohol can decrease butyrate (SCFA) production and administration of butyrate in the form of tributyrin mitigates alcohol-induced liver injury in mice (73).
Gut microbiome signature in alcohol dependence syndrome (ADS) is different than that in ALD/cirrhosis (ALC) (74). In a metagenomic analysis, patients with ADS and ALC have distinct species-level composition. Liver cirrhosis is characterized by a higher level of gut dysbiosis than alcoholism. Massive depletion of major commensals from the Bacteroidales order is accompanied by a rise of taxa normally inhabiting the oral cavity. The presence of oral species Lactobacillus salivarius, Veillonella parvula, and Streptococcus salivarius is more pronounced in ALC patients in comparison with both healthy controls as well as the ADS group. Interestingly, we found that Bifidobacterium abundance was significantly associated with the cirrhosis. ADS and ALC are associated with a different pool of Bifidobacterium and Lactobacillus species. While specific changes associated with alcoholic dependence suggest the onset of inflammatory environment in the gut, the hallmarks of the liver cirrhosis are likely linked to the impaired bile secretion.
In summary, there are some similarity and some unique differences in the gut microbiota composition in patients with NAFLD and ALD. This is illustrated in Figure 1.
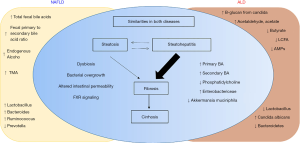
Circulating microbiome
The existence of microbial populations in “classically sterile” locations, including the blood, is a relatively new concept. The presence of bacteria-specific DNA in the blood has been reported in the literature for some time, yet the true origin of this is still the subject of much deliberation. Recently, Puri and colleagues show a significant increase in 16S copies/ng DNA in patients with AH (75). Circulating phylum Bacteroidetes is significantly decreased in both heavy drinking alcoholics (HDA) without liver disease, as well as with those with AH. In contrast, enrichment of circulating Fusobacteria is notable in all HDAs, with the greatest increase in HDA patients without liver disease compared to HDA with AH. Severe AH patients as expected have significantly higher endotoxemia. The predictive functional metagenomics suggest an enrichment of bacteria with genes related to methanogenesis and denitrification in HDAs patients and activation of a type III secretion system (linked to gram-negative bacterial virulence) in both HDAs without liver disease as well as those with severe AH. Metagenomics in severe AH predicts increased isoprenoid synthesis via mevalonate and anthranilate degradation, which are the known modulators of gram-positive bacterial growth and biofilm production, respectively.
Similarly, presence of liver fibrosis in patients with NAFLD is associated with a significantly higher blood 16S rDNA concentration than controls (76). Also, a significantly decreased relative abundance of Actinobacteria in the blood and markedly increased Proteobacteria characterizes obese NAFLD patients with liver fibrosis. Moreover, relative abundance in the blood for genera Sphingomonas, Bosea, and Variovorax significantly correlated with liver fibrosis. Together, circulating or blood microbiota signature offers a promising potential for disease biomarkers in NAFLD and ALD patient populations and deserve further investigations.
Fungal microbiome or mycobiome
Mycobiome and its role in chronic liver diseases is emerging. Mycobiome can be classified using fungal-specific internal transcribed spacer (ITS) amplicon sequencing (77,78). In the Human Microbiome Project (HMP) cohort, the Internal Transcribed Spacer 2 (ITS2) region and 18S rRNA gene methods are compared to interrogate human mycobiome (79). A greater resolution of the mycobiome membership is seen with ITS2 sequencing data compared to metagenomic and 18S rRNA gene sequencing data, suggesting that ITS2 is a more sensitive method for studying the mycobiome of stool samples. In contrast to bacterial communities, the human gut mycobiome is low in diversity and dominated by yeast including Saccharomyces, Malassezia, and Candida. Although several fungal species persist across most samples in the HMP cohort suggesting that a core human gut mycobiome exists, however a high inter- and intra-volunteer variability indicates that human gut mycobiome is not stable over time.
A recent study of chronic alcohol feeding in a murine model shows intestinal fungal overgrowth associated with translocation of fungal products as evident by elevated plasma levels of 1,3-β-D-glucan (80). Importantly, treatment with amphotericin B diminishes alcohol-associated increase in intestinal fungi and significantly lowers the plasma level of 1,3-β-D-glucan. In humans, when compared to the healthy controls, the richness and diversity of fungal species is decreased in alcoholic patients. Also, fecal Candida overgrowth characterizes patients with alcohol use, which is driven by increase in C. albicans. While Candida overgrowth is similar among patients with nonprogressive ALD, AH, and alcoholic cirrhosis; the average relative abundance of C. albicans is diminished with increasing severity of liver disease.
Serum anti–Saccharomyces cerevisiae IgG antibodies (ASCA) levels suggest systemic exposure and immune response to intestinal fungi. C. albicans is an important immunogen for ASCA and is likely the origin of an aberrant immune response in humans (81). Serum ASCA levels are significantly higher in patients with alcoholic cirrhosis compared to hepatitis B related cirrhosis or healthy controls. Moreover, in alcoholic cirrhosis patients with comparable MELD score, higher levels of ASCA (>8 U/mL) is associated with significantly higher five-year mortality which is more than doubled. Similarly, a significantly higher ASCA levels are seen in patients with AH compared to patients with AUD and to nonalcoholic controls (82). Moreover, ASCA levels (>34 IU/mL) significantly lower 90-day survival (59% vs. 80%) in patients with AH, with an adjusted hazard ratio of 3.13 (95% CI, 1.11–8.82; P=0.031). Altogether, patients with alcohol-associated liver disease have a lower fungal diversity and Candida overgrowth compared with controls. Notably, ASCA predicts survival in patients with alcoholic cirrhosis and AH. Intestinal mycobiome may therefore serve as an important therapeutic target to improve survival (82).
In a study of cirrhotics including patients with alcohol and NAFLD etiology, a new combined bacterial-fungal dysbiosis metric, with a higher Bacteroidetes/Ascomycota ratio, can independently predict 90-day hospitalizations (83). Moreover, antibiotics exposure in cirrhotics decreases bacterial and fungal diversity, which is associated with higher Candida and lower autochthonous (Ruminococcaceae, Lachnospiraceae, Clostridiales Cluster XIV) bacterial relative abundance. Proton pump inhibitors therapy in cirrhotics alters bacterial diversity and composition, however the mycobiota remained stable.
The proposed mechanism for mycobiome related liver damage in ALD is associated with increased systemic levels of β-1,3-glucan, a component of Candida. Increased β-1,3-glucan activates Kupffer cells in the liver to induce IL-1β production that in turn acts on hepatocytes to increase alcohol-induced inflammation, steatosis and hepatocyte injury (80).
Therapeutic perspectives
Part of the difficulty in developing effective treatments for NAFLD and ALD resides in the complexity in the disease pathogenesis and an effective model organism that can fully recapitulate human disease. Thus, there are currently no effective therapeutic interventions other than lifestyle modifications for the treatment of NAFLD and ALD. With our current understanding of the changes in the gut microbiome in NAFLD and ALD and the potential role that these organisms may have in liver disease progression, manipulating the human gut microbiome may provide a therapeutic target to ameliorate existing disease. A recent randomized, double-blinded, placebo study of cirrhotic patients demonstrated that Lactobacillus rhamnosus GG ATCC strain 53103 (LGG) could modulate systemic inflammation, fecal bile acids and the gut microbiome (84). It was observed that there was a significant reduction in inflammation through a decrease in serum endotoxins and TNFα. Another study showed that hospitalized patients admitted for alcohol treatment were given probiotics for 5 days (Bifidobacterium bifidum and Lactobacillus plantarum 8PA3) increased the levels of beneficial bacteria including Bifidobacteria and Lactobacilli (68). Results were further supported by a decrease in liver enzymes such as ALT, AST and GGT.
Fecal microbial transplant (FMT) is an emerging therapy for liver diseases with several ongoing clinical trials worldwide. Current FMT trials in NAFLD and ALD related liver disease are summarized in Table 1.
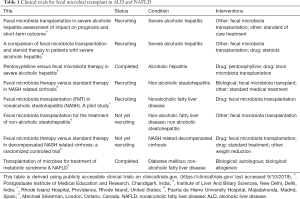
Full table
In summary, the gut microbiome is intricately balanced by various host factors and disruption could lead to liver disease and other various diseases. Recent studies have shed light on the importance of specific bacterial changes and bile acid dysregulation potentiating liver disease. Targeting bile acid-mediated signaling and the manipulation of the gut microbiota could potentially be a viable therapeutic target for various liver diseases.
General challenges in conducting and interpreting microbiome studies and its implications for NAFLD and ALD
Despite technical advances in the rapidly evolving field of microbiome research, the reproducibility and validity of findings deserve special attention. Large data sets with several competing variables often confound the observations compared to a rather clean disease state in an experimental model. In addition, complex ecosystem of host, genetics, and environmental factors such as the mode of delivery, diet, alcohol, medications, antibiotic use, etc. can cause variations in the composition as well as functionality of the gut microbiome. Another important but often overlooked technical aspect in human microbiome studies is the collection, processing and handling of the biospecimens. Another layer of complexity is the use of different sequencing methods, which yield different results. These include quantitative PCR, 16S rRNA sequencing, shotgun sequencing, metagenomics, etc. Also, different platforms for bioinformatics analyses, such as QIIME, Mothur and PICRUST, can introduce variability in the results. Further, measures of primary clinical endpoints also vary, i.e., liver biopsy, the so called gold standard, to non-invasive tools such as imaging or blood tests. Use of high-throughput DNA sequencing and powerful computational techniques have enabled us to interrogate the complexity of the microbiome, which also enhanced our understanding about the role of the microbiome in health and disease. Therefore, designing future studies in liver disease research should focus on overcoming some of these challenges to produce more robust, reproducible, generalizable and clinically applicable data at an individual and population level.
Acknowledgments
Funding: AA021179 (to P Puri) from National Institute on Alcohol Abuse and Alcoholism.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Ashwani K. Singal) for the series “Non-alcoholic Fatty Liver Disease and Alcoholic Liver Disease” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh.2020.02.18). The series “Non-alcoholic Fatty Liver Disease and Alcoholic Liver Disease” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Seki E, De Minicis S, Osterreicher CH, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 2007;13:1324-32. [Crossref] [PubMed]
- Isayama F, Hines IN, Kremer M, et al. LPS signaling enhances hepatic fibrogenesis caused by experimental cholestasis in mice. Am J Physiol Gastrointest Liver Physiol 2006;290:G1318-28. [Crossref] [PubMed]
- Chen YM, Liu Y, Zhou RF, et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep 2016;6:19076. [Crossref] [PubMed]
- Loomba G, Dhandapani M, Kaur S, et al. The Effectiveness of Personal Hygiene Practices on Non-Cuffed Central Vein Catheter-Related Infection in Patients Undergoing Hemodialysis: A Randomized Controlled Trial. Indian J Nephrol 2019;29:267-71. [Crossref] [PubMed]
- Nobili V, Mantovani A, Cianfarani S, et al. Prevalence of prediabetes and diabetes in children and adolescents with biopsy-proven non-alcoholic fatty liver disease. J Hepatol 2019;71:802-10. [Crossref] [PubMed]
- Kriz C, Flores S, Villarreal EG, et al. Impact of noonan syndrome on admissions for pediatric cardiac surgery. Minerva Pediatr 2019. [Epub ahead of print]. [PubMed]
- Schattenberg JM, Loomba R. Refining Noninvasive Diagnostics In Nonalcoholic Fatty Liver Disease: Closing the Gap to Detect Advanced Fibrosis. Hepatology 2019;69:934-6. [Crossref] [PubMed]
- Bashir MR, Wolfson T, Gamst AC, et al. Hepatic R2* is more strongly associated with proton density fat fraction than histologic liver iron scores in patients with nonalcoholic fatty liver disease. J Magn Reson Imaging 2019;49:1456-66. [Crossref] [PubMed]
- Tapper EB, Parikh ND. Mortality due to cirrhosis and liver cancer in the United States, 1999-2016: observational study. BMJ 2018;362:k2817. [Crossref] [PubMed]
- Bibbo S, Ianiro G, Dore MP, et al. Gut Microbiota as a Driver of Inflammation in Nonalcoholic Fatty Liver Disease. Mediators Inflamm 2018;2018:9321643.
- Hart CL, Morrison DS, Batty GD, et al. Effect of body mass index and alcohol consumption on liver disease: analysis of data from two prospective cohort studies. BMJ 2010;340:c1240. [Crossref] [PubMed]
- Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761-72. [Crossref] [PubMed]
- Hrubec Z, Omenn GS. Evidence of genetic predisposition to alcoholic cirrhosis and psychosis: twin concordances for alcoholism and its biological end points by zygosity among male veterans. Alcohol Clin Exp Res 1981;5:207-15. [Crossref] [PubMed]
- Whitfield JB. Meta-analysis of the effects of alcohol dehydrogenase genotype on alcohol dependence and alcoholic liver disease. Alcohol Alcohol 1997;32:613-9. [Crossref] [PubMed]
- Ambade A, Mandrekar P. Oxidative stress and inflammation: essential partners in alcoholic liver disease. Int J Hepatol 2012;2012:853175.
- Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology 2003;124:1488-99. [Crossref] [PubMed]
- Singh S, Osna NA, Kharbanda KK. Treatment options for alcoholic and non-alcoholic fatty liver disease: A review. World J Gastroenterol 2017;23:6549-70. [Crossref] [PubMed]
- Mutlu EA, Gillevet PM, Rangwala H, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol 2012;302:G966-78. [Crossref] [PubMed]
- Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology 2015;148:30-6. [Crossref] [PubMed]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol 2009;587:4153-8. [Crossref] [PubMed]
- Schnabl B, Brenner DA. Interactions Between the Intestinal Microbiome and Liver Diseases. Gastroenterology 2014;146:1513-24. [Crossref] [PubMed]
- Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018;555:210-5. [Crossref] [PubMed]
- Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105-8. [Crossref] [PubMed]
- Sampson TR, Debelius JW, Thron T, et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson's Disease. Cell 2016;167:1469-80.e12. [Crossref] [PubMed]
- Hughes HK, Rose D, Ashwood P. The Gut Microbiota and Dysbiosis in Autism Spectrum Disorders. Curr Neurol Neurosci Rep 2018;18:81. [Crossref] [PubMed]
- Ni J, Wu GD, Albenberg L, et al. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol 2017;14:573-84. [Crossref] [PubMed]
- Bajaj JS, Betrapally NS, Gillevet PM. Decompensated cirrhosis and microbiome interpretation. Nature 2015;525:E1-2. [Crossref] [PubMed]
- Hooks KB, O'Malley MA. Dysbiosis and Its Discontents. mBio 2017;8:e01492-17. [Crossref] [PubMed]
- Bajaj JS, Kakiyama G, Zhao D, et al. Continued Alcohol Misuse in Human Cirrhosis is Associated with an Impaired Gut-Liver Axis. Alcohol Clin Exp Res 2017;41:1857-65. [Crossref] [PubMed]
- Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60:940-7. [Crossref] [PubMed]
- Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology 2009;136:65-80. [Crossref] [PubMed]
- Ridlon JM, Harris SC, Bhowmik S, et al. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016;7:22-39. [Crossref] [PubMed]
- Nebe-von-Caron G, Stephens PJ, Hewitt CJ, et al. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J Microbiol Methods 2000;42:97-114. [Crossref] [PubMed]
- Ames NJ, Ranucci A, Moriyama B, et al. The Human Microbiome and Understanding the 16S rRNA Gene in Translational Nursing Science. Nurs Res 2017;66:184-97. [Crossref] [PubMed]
- Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220-30. [Crossref] [PubMed]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Jovel J, Patterson J, Wang W, et al. Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics. Front Microbiol 2016;7:459. [Crossref] [PubMed]
- Fukui H. Gut-liver axis in liver cirrhosis: How to manage leaky gut and endotoxemia. World J Hepatol 2015;7:425-42. [Crossref] [PubMed]
- Vancamelbeke M, Vermeire S. The intestinal barrier: a fundamental role in health and disease. Expert Rev Gastroenterol Hepatol 2017;11:821-34. [Crossref] [PubMed]
- Keshavarzian A, Holmes EW, Patel M, et al. Leaky gut in alcoholic cirrhosis: a possible mechanism for alcohol-induced liver damage. Am J Gastroenterol 1999;94:200-7. [Crossref] [PubMed]
- Enomoto N, Ikejima K, Yamashina S, et al. Kupffer cell sensitization by alcohol involves increased permeability to gut-derived endotoxin. Alcohol Clin Exp Res 2001;25:51S-4S. [Crossref] [PubMed]
- Kwong EK, Liu R, Zhao D, et al. The role of sphingosine kinase 2 in alcoholic liver disease. Dig Liver Dis 2019;51:1154-63. [Crossref] [PubMed]
- Doring B, Lutteke T, Geyer J, et al. The SLC10 carrier family: transport functions and molecular structure. Curr Top Membr 2012;70:105-68. [Crossref] [PubMed]
- Chiang JY. Bile acid metabolism and signaling. Compr Physiol 2013;3:1191-212. [PubMed]
- Kwong E, Li Y, Hylemon PB, et al. Bile acids and sphingosine-1-phosphate receptor 2 in hepatic lipid metabolism. Acta Pharm Sin B 2015;5:151-7. [Crossref] [PubMed]
- Kwong EK, Li X, Hylemon PB, et al. Sphingosine Kinases/Sphingosine 1-Phosphate Signaling in Hepatic Lipid Metabolism. Curr Pharmacol Rep 2017;3:176-83. [Crossref] [PubMed]
- Iracheta-Vellve A, Calenda CD, Petrasek J, et al. FXR and TGR5 Agonists Ameliorate Liver Injury, Steatosis, and Inflammation After Binge or Prolonged Alcohol Feeding in Mice. Hepatol Commun 2018;2:1379-91. [Crossref] [PubMed]
- Gabele E, Muhlbauer M, Dorn C, et al. Role of TLR9 in hepatic stellate cells and experimental liver fibrosis. Biochem Biophys Res Commun 2008;376:271-6. [Crossref] [PubMed]
- Hartmann P, Haimerl M, Mazagova M, et al. Toll-like receptor 2-mediated intestinal injury and enteric tumor necrosis factor receptor I contribute to liver fibrosis in mice. Gastroenterology 2012;143:1330-40.e1. [Crossref] [PubMed]
- Xie G, Zhong W, Li H, et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J 2013;27:3583-93. [Crossref] [PubMed]
- Bibbo S, Dore MP, Cammarota G. Response to: Comment on "Gut Microbiota as a Driver of Inflammation in Nonalcoholic Fatty Liver Disease". Mediators Inflamm 2018;2018:7328057.
- Bellentani S. The epidemiology of non-alcoholic fatty liver disease. Liver Int 2017;37 Suppl 1:81-4. [Crossref] [PubMed]
- Farrell GC. Signalling links in the liver: knitting SOCS with fat and inflammation. J Hepatol 2005;43:193-6. [Crossref] [PubMed]
- Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 2013;57:601-9. [Crossref] [PubMed]
- Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764-75. [Crossref] [PubMed]
- Loomba R, Seguritan V, Li W, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 2017;25:1054-62.e5. [Crossref] [PubMed]
- Caussy C, Tripathi A, Humphrey G, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun 2019;10:1406. [Crossref] [PubMed]
- Mouzaki M, Wang AY, Bandsma R, et al. Bile Acids and Dysbiosis in Non-Alcoholic Fatty Liver Disease. PLoS One 2016;11:e0151829. [Crossref] [PubMed]
- Arendt BM, Ma DW, Simons B, et al. Nonalcoholic fatty liver disease is associated with lower hepatic and erythrocyte ratios of phosphatidylcholine to phosphatidylethanolamine. Appl Physiol Nutr Metab 2013;38:334-40. [Crossref] [PubMed]
- Puri P, Baillie RA, Wiest MM, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology 2007;46:1081-90. [Crossref] [PubMed]
- Corbin KD, Zeisel SH. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr Opin Gastroenterol 2012;28:159-65. [Crossref] [PubMed]
- Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011;472:57-63. [Crossref] [PubMed]
- Maher JJ. New insights from rodent models of fatty liver disease. Antioxid Redox Signal 2011;15:535-50. [Crossref] [PubMed]
- Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2013;62:1787-94. [Crossref] [PubMed]
- Bode JC, Bode C, Heidelbach R, et al. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology 1984;31:30-4. [PubMed]
- Yan AW, Fouts DE, Brandl J, et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology 2011;53:96-105. [Crossref] [PubMed]
- Llopis M, Cassard AM, Wrzosek L, et al. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut 2016;65:830-9. [Crossref] [PubMed]
- Kirpich IA, Solovieva NV, Leikhter SN, et al. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol 2008;42:675-82. [Crossref] [PubMed]
- Leclercq S, Matamoros S, Cani PD, et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A 2014;111:E4485-93. [Crossref] [PubMed]
- Axelson M, Mork B, Sjovall J. Ethanol has an acute effect on bile acid biosynthesis in man. FEBS Lett 1991;281:155-9. [Crossref] [PubMed]
- Kakiyama G, Hylemon PB, Zhou H, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol 2014;306:G929-37. [Crossref] [PubMed]
- Chen P, Torralba M, Tan J, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology 2015;148:203-14.e16. [Crossref] [PubMed]
- Cresci GA, Glueck B, McMullen MR, et al. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol 2017;32:1587-97. [Crossref] [PubMed]
- Dubinkina VB, Tyakht AV, Odintsova VY, et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome 2017;5:141. [Crossref] [PubMed]
- Puri P, Liangpunsakul S, Christensen JE, et al. The circulating microbiome signature and inferred functional metagenomics in alcoholic hepatitis. Hepatology 2018;67:1284-302. [Crossref] [PubMed]
- Lelouvier B, Servant F, Paisse S, et al. Changes in blood microbiota profiles associated with liver fibrosis in obese patients: A pilot analysis. Hepatology 2016;64:2015-27. [Crossref] [PubMed]
- Fouts DE, Szpakowski S, Purushe J, et al. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One 2012;7:e48289. [Crossref] [PubMed]
- Bokulich NA, Mills DA. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl Environ Microbiol 2013;79:2519-26. [Crossref] [PubMed]
- Nash AK, Auchtung TA, Wong MC, et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017;5:153. [Crossref] [PubMed]
- Yang AM, Inamine T, Hochrath K, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest 2017;127:2829-41. [Crossref] [PubMed]
- Standaert-Vitse A, Jouault T, Vandewalle P, et al. Candida albicans is an immunogen for anti-Saccharomyces cerevisiae antibody markers of Crohn's disease. Gastroenterology 2006;130:1764-75. [Crossref] [PubMed]
- Lang S, Duan Y, Liu J, et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology 2020;71:522-38. [Crossref] [PubMed]
- Bajaj JS, Liu EJ, Kheradman R, et al. Fungal dysbiosis in cirrhosis. Gut 2018;67:1146-54. [Crossref] [PubMed]
- Bajaj JS, Heuman DM, Hylemon PB, et al. Randomised clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther 2014;39:1113-25. [Crossref] [PubMed]
Cite this article as: Kwong EK, Puri P. Gut microbiome changes in Nonalcoholic fatty liver disease & alcoholic liver disease. Transl Gastroenterol Hepatol 2021;6:3.


