
Video capsule endoscopy: pushing the boundaries with software technology
Introduction
Since its introduction in 2000, the Video Capsule Endoscopy (VCE) has transformed imaging of the small bowel (1). VCE offers a non-invasive and well tolerated modality for examining the entire small intestinal mucosa. The capsule is swallowed and passes passively through the gastrointestinal (GI) tract while acquiring images, which need to be reviewed at a later stage. It has become an established modality for the investigation of obscure iron deficiency anemia and occult GI bleeding, and can be used for the diagnosis and assessment of Crohn’s disease and polyposis syndromes (2-4). The development of device-assisted enteroscopy allows small intestinal lesions detected on VCE to be subsequently treated endoscopically.
It is nearly 20 years since VCE was first introduced and the way in which we read VCE has not changed significantly. Reading of examination is still very time intensive and prone to reader error. A VCE recording may be several hours in length with tens of thousands of frames but the causative pathologic lesion may be seen in only a few frames. This can test the limits of human concentration, increasing the possibility of missed lesions. This review highlights the advances in software technology, and particularly the recent advances in artificial intelligence (AI), which aim to address these challenges.
Capsule endoscopy systems
In order to appreciate efforts to enhance software, it is important to outline the available hardware. There are five commercially available small bowel capsule endoscopy (SBCE) systems (5), each with the same core components. This includes an imaging device, lens, a light source and a battery, all within a plastic casing resistant to digestive juices. The batteries give a lifespan of 10–15 hours, enabling over 5,000 images to be acquired at a rate of 2–6 frames per second (fps). In most cases, images captured by the capsule are wirelessly transmitted to a receiving device attached to a belt worn by the patient, except in the case of CapsoCam, where the data is stored within the capsule and the capsule needs to be retrieved after the examination. The images are downloaded onto a computer and read using proprietary software. A comparison of the different systems is summarized in Table 1. Several trials have compared the performance of one capsule system with another (3). These have mostly compared PillCam with MiroCam or Endocapsule, and have not detected any significant differences between the different systems.
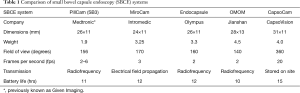
Full table
Conventional capsule interpretation
Within the various VCE software programs, adjustments can be made to the number of frames viewed and the speed at which frames are presented. Frames can be read as either single view (SV), dual view (DV) or quad view (QV), as either sequential or overlapping images. For example, the QV overlap mode has each image moving across the screen in four frames. This requires a greater use of peripheral vision compared to a single central image.
Increasing the speed at which frames are presented, expressed as fps, results in an increased chance of missing lesions. This was demonstrated in a study with a single 15-minute video clip containing 60 frames with different frame views and rates (6), where the best setting for lesion detection was QV overlapping at 10 fps. A further study showed that the diagnostic yield of SV was 26% at 25 fps and 45% at 15 fps, whilst QV at speeds up to 30 fps had no reduction in accuracy (7).
The European Society of Gastrointestinal Endoscopy (ESGE) technical review on capsule endoscopy suggest the optimal reading speed is a maximum of 10 fps in an SV mode or up to 20 fps in dual or QV modes (8); as well as reducing reading speeds in the proximal small bowel. This is because the passage of a capsule is faster here than the ileum, which is demonstrated by the ampulla, the only landmark in the small bowel, being seen in only 10% of SBCE examinations (9).
Software enhancements
VCE would seem to be an ideal modality for capitalizing on advances in information technology (10), aiming to aide in both enhancing lesion detection and reducing the number of normal images reviewed.
Suspected blood indicator (SBI)
The SBI is a rapid viewing tool that can be activated within the various capsule software programs. It aims to identify bleeding lesions by highlighting frames with an excess number of red pixels. When the indication for the VCE is gastrointestinal bleeding or iron deficiency anemia, this would allow the reader to rapidly identify the lesion and location without having to review the entire capsule examination. A meta-analysis of 16 studies, including 2,049 patients, showed a sensitivity of 98.8% in the detection of actively bleeding lesions. However, for detection of lesions with the potential to bleed but were not actively bleeding, the sensitivity was only 55.3% and specificity 57.8% (11). The poor accuracy of the SBI function means it cannot be used as a time-saving technique in clinical practice but may be used to ensure lesions have not been missed after the initial reading.
Image selection
The passage of the capsule is passive as it is dictated by bowel peristalsis, leading to multiple duplicate images being captured. Removal of these images with only clinically relevant images would dramatically reduce reading times. There have been several software algorithms developed for this purpose.
The most studied is QuickView, an automated fast viewing mode available on Rapid software (Medtronic, Dublin, Ireland). The proportion of images excluded can be determined by the reader, thus significantly reducing reading time. Unfortunately, performance is poor, with a lesion miss rate ranging from 6.5% to 12% (12-14), although it does have reasonable accuracy in the detection of major lesions. Most of the missed lesions were single isolated lesions, including polyps and isolated vascular or inflammatory lesions. A limitation of these studies is that there is significant inter-observer variability on interpretations of capsule endoscopies, even between experts (15,16).
The utility of QuickView is further undermined given that reading in SV or DV at 20 fps was shown to be more accurate, although not as rapid (17). Another study showed that simply covering half the screen with a piece of paper when viewing in QD sequential resulted in a lower lesion miss rate (13). This implies that the random exclusion of half the images was more accurate than the selection of excluded frames by QuickView. Despite this, there are certain clinical situations, such as overt obscure gastrointestinal bleeding, where QuickView can be used effectively (18).
Other rapid-viewing algorithms have less evidence. The OMOM similar picture elimination software has three modes that allow images to be removed and improve reading times. Only one of the modes showed a sensitivity >85% and this was associated with a modest time saving of 9 min (19). EndoCapsule has a similar software with two modes: express selected removes repeated images and auto adjust maintains the repeated images but at an increased speed within the viewing stream. This had an accuracy of 97.5%. The more recent Omni mode software reduces images displayed by 65% through the ‘intelligent’ removal of repeated and overlapping images. A multicenter trial from Japan with 40 selected cases showed this software was able to correctly remove images while keeping all the pre-identified major lesions (20).
Lesion characterization
Advanced imaging modalities are becoming more commonly used in endoscopic detection and characterization. Fuji Intelligent Color Enhancement (FICE) (Fujifilm, Saitama, Japan) is a post-processing visual enhancement technology that enhances mucosal surface patterns by using software to convert white light images to a restricted range of wavelengths (21). When FICE is used with PillCam, the results have been disappointing with no improvement in lesion detection (22), although enhancements in lesion delineation were shown. This is likely to be due to the passive movement of the VCE compared to flexible endoscopy.
Another image enhancement is the Blue Mode (BM) modality for PillCam, which can be used alone or in conjunction with FICE. Findings for BM have been mixed, with one study finding an increased detection rate (23) whilst another found no difference in detection rate and no benefit over conventional white light for delineating small bowel lesions (24).
Three-dimensional representation
VCE recordings are two dimensional, and it has been hypothesized that diagnosis may be hampered by the lack of appreciation of visual depth. Efforts have been made to reconstruct three dimensional surfaces from two dimensional images so as to improve diagnostic evaluation of lesions seen. One technique is to use algorithms that estimate depth from shading (25). Karargyris et al. compared four publicly available Shape from Shading (SfS) algorithms and found the Tsai’s SfS algorithm to perform better than other SfS algorithms (26). However, the clinical utility of this is still uncertain.
AI
AI refers to any technique that enables computers to mimic cognitive function displayed by humans, such as learning and problem solving. Machine learning (ML) is a subset of AI that includes statistical techniques allowing the automatic detection of data patterns. These are used to predict future data or enable decision making. ML is driven by large amounts of data and algorithms rather than inputting specific codes.
Deep learning (DL) is a subset and more recent development of ML. It is the process by which a computer collects, analyses and processes the data it needs without receiving explicit instructions. It is therefore characterized by self-learning, as the program itself extracts key features from a data set. DL is developed in an artificial neural network (ANN), which uses data input and output hierarchies in a similar fashion to brain neurons to learn data. The strength of the system therefore relies on the strength of the underlying database. Convolutional neural network (CNN) are a type of ANN based on principles of the visual cortex of the brain in processing and recognizing images.
In medicine, AI has the potential to both improve clinical care and expedite clinical processes, thus relieving the burden on medical professionals. Already, AI has successfully been used to improve performance in medical diagnostics, including the diagnosis of diabetic retinopathy and cutaneous malignancies (27). In endoscopy, AI is being investigated to enhance computer-aided detection (CADe) and computer aided diagnosis (CADx) (28). The potential of CADe is demonstrated in an open label trial recently reported by Wang et al. (29). They randomized 1,058 patients undergoing colonoscopy: assistance from the real time automatic polyp detection system resulted in an adenoma detection rate of 29.1% compared to 20.3% in those randomized to conventional colonoscopy. In a separate study, the potential of CADx was shown by a DL method being able to differentiate neoplastic from hyperplastic diminutive polyps with a high degree of accuracy (30).
For VCE, there is great potential for AI to improve the time-intensive nature of reviewing examinations, particularly given the lack of success of existing software technology (31). In addition, the CAD systems are not subject inter- and intra-observer variance and fatigue, which are common in human readings. However, there are challenges with CNN-based diagnostic programs for capsule endoscopy. Images have low resolution due to constraints imposed by the hardware, with multiple orientations due to the free motion of the capsule, as well as obscuring artefacts such as bile and fecal material. Despite this, studies have progressed rapidly and show promise for clinical application in the near future.
Detection of single aspects or lesions
Several pilot studies have demonstrated the potential of DL for VCE but mostly concentrating on a single aspect or lesion for detection. Zou et al. used a CNN based algorithm to differentiate images into organs of origin: stomach, small bowel and colon (32). This achieved an accuracy of 95.52% for 15,000 images from 25 patients. Seguí et al. developed a CNN system to characterize small intestine motility events, including turbid, bubbles, clear blob and wrinkle (33). This demonstrated a classification accuracy of 96%, which outperformed other classifiers by 14%. This is important to demonstrate, as capsule reading is hindered by such intraluminal contents, so any DL approach would need to distinguish these from pathology.
For detecting gastrointestinal bleeding, several studies have used DL based approaches to achieve sensitivities and specificities of up to 100% (34-36). These results probably owe to the obvious difference in color hue of blood compared to more subtle changes looked at in other studies. They offer the potential of rapid and effective identification of bleeding, although it is unclear if there is any significant advantage over the SBI, which has similarly high sensitivity for actively bleeding lesions.
For the detection of polyps, Yuan and Meng used a DL based approach to achieve an accuracy of 98% for 4,000 images from 35 patients and outperforming existing polyp recognition methods (37).
A CNN developed using capsule endoscopy images from patients with and without celiac disease (38) reported a sensitivity and specificity of 100% when validated with a with a set of 10 patients. This compares with previously reported low sensitivity of visual diagnosis of celiac disease by VCE (39). Interestingly, the evaluation confidence of the algorithm correlated with the severity of villous atrophy in the small bowel. This may have application for those patients unwilling or unable to undertake a diagnostic esophagogastroduodenoscopy for histological diagnosis.
A CNN developed to detect hookworms (40) was tested on 440,000 images from 11 patients. This resulted in 88.5% accuracy and 84.6% sensitivity for hookworm detection. However, it is unclear whether this would have much clinical application, since the majority of the population affected by hookworm are from resource-limited countries and diagnosis is usually achieved simply from by identifying ova in the stool.
More recently, Leenhardt et al. used a CNN to detect GI angioectasias in the small bowel (41). They used a French national database for VCE incorporating multiple centers, with still frames from 4,166 PillCam SB3 examinations and 2,946 still frames showing vascular lesions. The same number of normal still frames were used as the control dataset. They achieved a sensitivity of 100% for the detection of GI angioectasias with a specificity of 96%, with a reading time of approximately 39 minutes for a full-length capsule study.
Detection of multiple types of lesions
In real clinical situations, there may be multiple possible causes that need to be looked for. For example, iron deficiency anemia could be due to lesions from Crohn’s disease, coeliac disease, angioectasias and so on. Therefore, any DL algorithm would need to identify multiple possible lesions to be clinically applicable.
A recent study by Ding et al. demonstrated the feasibility of identifying and discriminating multiple possible lesions. Their CNN based algorithm was developed with 113,426,569 images from 6,970 patients from multiple centers (42). Images were classified as normal, lymphangiectasia, lymphatic follicular hyperplasia, vascular disease, inflammation, ulcer, bleeding, polyps, protruding lesion, diverticulum, parasite or other. The algorithm identified abnormalities with 99.9% sensitivities and a reading time of 5.9 minutes, whilst gastroenterologists using conventional reading had much lower sensitivities and of course much longer reading times.
Conclusions
Advances in software technology to aid the reading time and detection rate of lesions have been disappointing until the advent of DL as a form of AI. This promises to revolutionize capsule endoscopy reading. There are still challenges ahead, with the need for robust validation and importantly, the issue of medicolegal responsibility. However, we will inevitably see this in clinical practice where is will greatly enhance both the speed and accuracy of diagnosis.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Krish Ragunath, Philip WY Chiu) for the series “Advanced Endoscopic Imaging of the GI Tract” published in Translational Gastroenterology and Hepatology. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tgh.2020.02.01). The series “Advanced Endoscopic Imaging of the GI Tract” was commissioned by the editorial office without any funding or sponsorship. SB: research funding from Olympus. FP has no conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Iddan G, Meron G, Glukhovsky A, et al. Wireless capsule endoscopy. Nature 2000;405:417. [Crossref] [PubMed]
- Pennazio M, Spada C, Eliakim R, et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small-bowel disorders: European Society of Gastrointestinal Endoscopy (ESGE) clinical guideline. Endoscopy 2015;47:352-76. [Crossref] [PubMed]
- Koulaouzidis A, Rondonotti E, Karargyris A.. Small-bowel capsule endoscopy: a ten-point contemporary review. World J Gastroenterol 2013;19:3726. [Crossref] [PubMed]
- McAlindon ME, Ching HL, Yung D, et al. Capsule endoscopy of the small bowel. Ann Transl Med 2016;4:369. [Crossref] [PubMed]
- Beg S, Parra-Blanco A, Ragunath K. Optimising the performance and interpretation of small bowel capsule endoscopy. Frontline Gastroenterol 2018;9:300-8. [Crossref] [PubMed]
- Nakamura M, Murino A, O'Rourke A, et al. A critical analysis of the effect of view mode and frame rate on reading time and lesion detection during capsule endoscopy. Dig Dis Sci 2015;60:1743-7. [Crossref] [PubMed]
- Zheng Y, Hawkins L, Wolff J, et al. Detection of lesions during capsule endoscopy: physician performance is disappointing. Am J Gastroenterol 2012;107:554-60. [Crossref] [PubMed]
- Rondonotti E, Spada C, Adler S, et al. Small-bowel capsule endoscopy and device-assisted enteroscopy for diagnosis and treatment of small bowel disorders: ESGE Technical Review. Endoscopy 2018;50:423-46. [Crossref] [PubMed]
- Koulaouzidis A, Plevris JN. Detection of the ampulla of Vater in small bowel capsule endoscopy: experience with two different systems. J Dig Dis 2012;13:621-7. [Crossref] [PubMed]
- Iakovidis DK, Koulaouzidis A. Software for enhanced video capsule endoscopy: challenges for essential progress. Nat Rev Gastroenterol Hepatol 2015;12:172-86. [Crossref] [PubMed]
- Yung DE, Sykes C, Koulaouzidis A. The validity of suspected blood indicator software in capsule endoscopy: a systematic review and meta-analysis. Expert Rev Gastroenterol Hepatol 2017;11:43-51. [Crossref] [PubMed]
- Hosoe N, Rey JF, Imaeda H, et al. Evaluations of capsule endoscopy software in reducing the reading time and the rate of false negatives by inexperienced endoscopists. Clin Res Hepatol Gastroenterol 2012;36:66-71. [Crossref] [PubMed]
- Saurin JC, Lapalus MG, Cholet F, et al. Can we shorten the small bowel capsule reading time with the “Quick-view” image detection system? Dig Liver Dis 2012;44:477-81. [Crossref] [PubMed]
- Westerhof J, Koornstra JJ, Weersma RK. Can we reduce capsule endoscopy reading times? Gastrointest Endosc 2009;69:497-502. [Crossref] [PubMed]
- Lai LH, Wong GL, Chow DK, et al. Inter-observer variations on interpretation of capsule endoscopies. Eur J Gastroenterol Hepatol 2006;18:283-6. [Crossref] [PubMed]
- Jang BI, Lee SH, Moon JS, et al. Inter-observer agreement on the interpretation of capsule endoscopy findings based on capsule endoscopy structured terminology: a multicenter study by the Korean Gut Image Study Group. Scand J Gastroenterol 2010;45:370-4. [Crossref] [PubMed]
- Kyriakos N, Karagiannis S, Galanis P, et al. Evaluation of four time-saving methods of reading capsule endoscopy videos. Eur J Gastroenterol Hepatol 2012;24:1276-80. [Crossref] [PubMed]
- Koulaouzidis A, Smirnidis A, Douglas S, et al. QuickView in small-bowel capsule endoscopy is useful in certain clinical settings, but QuickView with Blue Mode is of no additional benefit. Eur J Gastroenterol Hepatol 2012;24:1099-104. [Crossref] [PubMed]
- Subramanian V, Mannath J, Telakis E, et al. Efficacy of new playback functions at reducing small-bowel wireless capsule endoscopy reading times. Dig Dis Sci 2012;57:1624-8. [Crossref] [PubMed]
- Hosoe N, Watanabe K, Miyazaki T, et al. Evaluation of performance of the Omni mode for detecting video capsule endoscopy images: A multicenter randomized controlled trial. Endosc Int Open 2016;4:E878-82. [Crossref] [PubMed]
- Osawa H, Yamamoto H. Present and future status of flexible spectral imaging color enhancement and blue laser imaging technology. Dig Endosc 2014;26:105-15. [Crossref] [PubMed]
- Yung DE, Boal Carvalho P, Giannakou A, et al. Clinical validity of flexible spectral imaging color enhancement (FICE) in small-bowel capsule endoscopy: a systematic review and metaanalysis. Endoscopy 2017;49:258-69. [Crossref] [PubMed]
- Abdelaal UM, Morita E, Nouda S, et al. Blue mode imaging may improve the detection and visualization of small bowel lesions: A capsule endoscopy study. Saudi J Gastroenterol 2015;21:418-22. [Crossref] [PubMed]
- Koulaouzidis A, Douglas S, Plevris JN. Blue mode does not offer any benefit over white light when calculating Lewis score in small-bowel capsule endoscopy. World J Gastrointest Endosc 2012;4:33-7. [Crossref] [PubMed]
- Koulaouzidis A, Karargyris A, Rondonotti E, et al. Three-dimensional representation software as image enhancement tool in small-bowel capsule endoscopy: a feasibility study. Dig Liver Dis 2013;45:909-14. [Crossref] [PubMed]
- Karargyris A, Rondonotti E, Mandelli G, et al. Evaluation of 4 three-dimensional representation algorithms in capsule endoscopy images. World J Gastroenterol 2013;19:8028-33. [Crossref] [PubMed]
- Yang YJ, Bang CS. Application of artificial intelligence in gastroenterology. World J Gastroenterol 2019;25:1666-83. [Crossref] [PubMed]
- Alagappan M, Glissen Brown JR, Mori Y, et al. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc 2018;10:239-49. [Crossref] [PubMed]
- Wang P, Berzin TM, Glissen Brown JR, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut 2019;68:1813-9. [Crossref] [PubMed]
- Chen PJ, Lin MC, Lai MJ, et al. Accurate Classification of Diminutive Colorectal Polyps Using Computer-Aided Analysis. Gastroenterology 2018;154:568-75. [Crossref] [PubMed]
- Hwang Y, Park J, Lim YJ, et al. Application of Artificial Intelligence in Capsule Endoscopy: Where Are We Now? Clin Endosc 2018;51:547-51. [Crossref] [PubMed]
- Zou Y, Li L, Wang Y, et al. Classifying digestive organs in wireless capsule endoscopy images based o deep convolutional neural network. Singapore: 2015 IEEE International Conference of Digital Signal Processing (DSP), 2015:1274-8.
- Seguí S, Drozdzal M, Pascual G, et al. Generic feature learning for wireless capsule endoscopy analysis. Comput Biol Med 2016;79:163-72. [Crossref] [PubMed]
- Hassan AR, Haque MA. Computer-aided gastrointestinal hemorrhage detection in wireless capsule endoscopy videos. Comput Methods Programs Biomed 2015;122:341-53. [Crossref] [PubMed]
- Jia Xiao, Meng MQ. A deep convolutional neural network for bleeding detection in Wireless Capsule Endoscopy images. Conf Proc IEEE Eng Med Biol Soc 2016;2016:639-42. [Crossref] [PubMed]
- Li P, Li Z, Gao F et al. Convolutional neural networks for intestinal haemorrhage detection in wireless capsule endoscopy images. Hong Kong: 2017 IEEE International Conference on Multimedia and Expo (ICME), 2017:1518-23.
- Yuan Y, Meng MQ. Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 2017;44:1379-89. [Crossref] [PubMed]
- Zhou T, Han G, Li BN, et al. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med 2017;85:1-6. [Crossref] [PubMed]
- Petroniene R, Dubcenco E, Baker JP, et al. Given capsule endoscopy in celiac disease: evaluation of diagnostic accuracy and interobserver agreement. Am J Gastroenterol 2005;100:685-94. [Crossref] [PubMed]
- He JY, Wu X, Jiang YG, et al. Hookworm detection in Wireless Capsule Endoscopy images with deep learning. IEEE Trans Image Process 2018;27:2379-92. [Crossref] [PubMed]
- Leenhardt R, Vasseur P, Li C, et al. A neural network algorithm for detection of GI angiectasia during small-bowel capsule endoscopy. Gastrointest Endosc 2019;89:189-94. [Crossref] [PubMed]
- Ding Z, Shi H, Zhang H, et al. Gastroenterologist-level Identification of Small Bowel Diseases and Normal Variants by Capsule Endoscopy Using a Deep-learning Model. Gastroenterol 2019. [Epub ahead of print].
Cite this article as: Phillips F, Beg S. Video capsule endoscopy: pushing the boundaries with software technology. Transl Gastroenterol Hepatol 2021;6:17.


