
To BE or not to BE: non-invasive screening for Barrett’s esophagus, dysplasia and adenocarcinoma
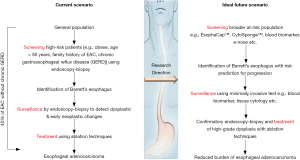
The EsophaCap™ DNA methylation biomarker validation study by Wang, Kambhampati and colleagues (1) follows a spade of reports (summarized in Table 1) aiming to develop screening tests for Barrett’s esophagus (BE). BE is a non-malignant metaplastic condition with little or no clinical symptoms. Arguably, the main importance of diagnosing BE is the increased risk of the patient to progress to esophageal adenocarcinoma (EAC). EAC has surpassed esophageal squamous cell carcinoma as the most common histological type of esophageal cancer in Western countries and its incidence is predicted to grow, possibly associated with life style and obesity (6,7). The majority of EAC are diagnosed at late stages, when it is associated with poor survival (6). Early detection and eradication at dysplasia stages is associated with improved survival (8). To detect early dysplastic changes, BE patients routinely undergo endoscopic surveillance, but endoscopic screening is currently recommended only for patients with multiple risk factors (Figure 1), due to the invasive nature the procedure (9,10).

Full table
Although newer techniques such as transnasal endoscopy (11,12) and capsule endoscopy (13-15) do not require patient sedation, they are still considered invasive. A less invasive biomarker-based test with high sensitivity and specificity in detecting early EAC or dysplasia will be a game changer in the diagnosis and patient outcomes for this aggressive cancer. Therefore, recent research efforts have focussed on molecular biomarkers for less invasive tests for BE screening and surveillance. As summarized in Table 1, Cytosponge™ (3) and inflatable balloon (2), and now EsophaCap™ (1,5) have been tested as alternatives for esophagogastroduodenoscopy, with favourable patient preference for the non-endoscopic procedures, and high completion rates (2,3,5). Unfortunately, different methylation biomarker candidates were measured, hence the performance between modes of sampling and diagnostic value of biomarker candidates/panels could not be separated.
Biomarker development involves multiple phases, broadly termed discovery, qualification and (multiple) validation. Not all biomarker candidates are expected to pass through all stages. Apart from biological differences of the cohorts, changes in sample type and/or technical pipelines can contribute to biomarkers failing to validate between phases. The current paper by Wang et al. (1) followed up eight methylation biomarkers previously reported by this team as tissue-based biomarkers for BE and BE progression to high-grade dysplasia/EAC. In this study, the group used EsophaCap™-based cytology sampling to evaluate biomarker performance for BE detection. While the cohort size is relatively modest, this is the only study that evaluated statistical model in a separate test set (see Table 1).
Although the current study (1), and the 2018 study by Iyer et al. (5) both evaluated the EsophaCap™ sponge device for cytological sampling, there are many differences. Apart from the different biomarker panels, Iyer et al. reported 38.0 and 30.8 µg of DNA yield using 100 and 20 ppi sponge (5) while the DNA yield in the current study is 5.7 µg (1). Furthermore, Iyer et al. (5) reported correlation between BE length and methylation levels of target genes while Wang et al. did not report analysis of BE length (1). As cytological sponge sampling is a low yield procedure without image guidance, it is important to know if the correlation exists between length of BE tissue and levels of biomarker candidate. A previous study by Chettouh et al. (4) also reported correlation of cytology methylation biomarkers with BE length.
A variety of statistical approaches were applied by Wang et al. (1) to develop biomarker panels. Age was included as a variable in all multimarker models, however, as both training and test cohorts showed significant difference in age of the control and BE groups, this may have been confounding factor. In the test set, age alone could predict BE with 80% accuracy. This is probably why the AUROC is higher in the test set than in the training set. It would have been interesting to understand how much of the predictive power of the model is due to the markers alone, without age in the model. Indeed, methylation of genes are reported to alter with age (4), hence this aspect should be re-evaluated. Another unusual feature of the prospective cohort reported by Wang et al. (1) is the high proportion of BE compared to the literature due to gastrointestinal symptoms described in cases. It is plausible that ability of biomarker candidates tested in this cohort will differ in any future validation studies in independent cohorts with lower rates (i.e., more realistic rates) of BE diagnosis.
While evaluating different types of omics data, it is necessary to consider the mathematical properties of each omics data. For example, the DNA methylation values presented by Wang et al. (1) tend to be skewed [see Figure 3 of Wang et al. (1)]. The asymmetric data distribution affects the Lasso regression models, similarly to traditional logistic regression. Considering the data distribution, perhaps the most skewed markers would have performed better if they had been log transformed. Furthermore, Lasso regression tends to select a single variable from groups of correlated variables and so potentially predictive markers can be interpreted as not predictive due to their correlation to other markers in the set. Hence, it would have been interesting also to see the correlations between the markers, including demographic variable. Presentation of dichotomized demographic data [as means, standard errors and P values in Table S2 of Wang et al. (1)] are actually not meaningful for highly skewed data.
As a large proportion of EAC cases arises with no prior diagnosis of BE or chronic reflux (16), non-invasive screening in broader population is needed to reduce EAC burden. While it is encouraging to see much activity in non-invasive diagnosis of BE. However, the true utility of screening and increasing diagnosis of BE needs to be considered. In contrast to colon cancer where adenoma (dysplasia) detection and therapy can be performed at same setting, screening early dysplastic Barrett’s and halting progression to high-grade dysplasia and adenocarcinoma is not as simple. As currently general treatment is not recommended for non-dysplastic BE, a diagnosis of BE can lead to anxiety and potentially overtreatment (17). Moreover, overdiagnosis of BE will result into larger patient pool to be surveilled using endoscopic techniques which may lead to increased economic burden on the healthcare system. Therefore, a more effective goal would be screening for dysplasia or EAC in patients having several risk factors for EAC. Risk prediction models could be used for such a directed screening strategy (Figure 1). Indeed, the BETS2 trial used the Cytosponge™ device with a biomarker panel to derive a risk prediction model consisting of glandular atypia, P53 abnormality, and Aurora kinase A positivity, age, waist-to-hip ratio, and length of the BE segment (18). The latter parameter requires endoscopic investigation though.
For even less invasive sampling, blood biomarkers as surrogates are an attractive option. To this end, our group has validated serum glycoprotein biomarkers that may be useful for detecting high-grade dysplasia and EAC from non-dysplastic BE and BE with low-grade dysplasia (19). Compared to cytology and transnasal endoscopy, blood biomarkers are less costly and can be easily conducted at primary care sites. In addition to proteins, miRNA changes in the serum related to dysplasia/EAC have also shown promise as surrogate biomarkers (20). Last but not the least, Chan and colleagues compared ‘breath print’ of BE and non-BE individuals using e-nose device and could differentiate BE from normal individuals with AUROC of 0.79 (21). All of these candidate biomarkers require multi-cohort validation including early dysplastic condition with longitudinal study design. With emergence of novel technologies, more than one screening/surveillance modality may be available to clinicians for selection to tailor for individual patient management. Such multiple ways of patient screening/monitoring are already available for colon cancer (e.g., colonoscopy, annual fecal immunochemical test, or DNA based testing). However, this translational journey for BE detection/monitoring from bench-to-bedside will be long and require rigorous clinical validation.
In conclusion, recent studies report promising results in molecular markers for non-invasive diagnosis of BE, and/or EAC. Due to the overall low incidence rate of dysplasia diagnosis and progression to EAC, multi-site collaborative studies are needed to fully evaluate the clinical utility of these markers. Critical evaluation of the clinical pathways should be conducted, to ensure the trials address actionable clinical needs and not lead to overdiagnosis and overtreatment. If an appropriate point-of-care tool could be standardized to detect at-risk BE patients, subsequently an intensive surveillance protocol for this risk group with enhanced imaging guided endoscopy (22) could be applied to detect dysplasia and treat these patients using ablative modalities. With development and implementation of non-invasive tests and risk prediction algorithms, the ideal scenario for EAC prevention through screening (Figure 1) may become a future reality.
Acknowledgements
We thank Madeleine Flynn for artwork.
Footnote
Conflicts of Interest: AK Shah and MM Hill are inventors on patent applications for serum glycoprotein biomarkers for esophageal adenocarcinoma. The other authors have no conflicts of interest to declare.
References
- Wang Z, Kambhampati S, Cheng Y, et al. Methylation Biomarker Panel Performance in EsophaCap Cytology Samples for Diagnosing Barrett's Esophagus: A Prospective Validation Study. Clin Cancer Res 2019;25:2127-35. [Crossref] [PubMed]
- Moinova HR, LaFramboise T, Lutterbaugh JD, et al. Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett's esophagus. Sci Transl Med 2018. [Crossref] [PubMed]
- Ross-Innes CS, Debiram-Beecham I, O'Donovan M, et al. Evaluation of a minimally invasive cell sampling device coupled with assessment of trefoil factor 3 expression for diagnosing Barrett's esophagus: a multi-center case-control study. PLoS Med 2015;12:e1001780. [Crossref] [PubMed]
- Chettouh H, Mowforth O, Galeano-Dalmau N, et al. Methylation panel is a diagnostic biomarker for Barrett's oesophagus in endoscopic biopsies and non-endoscopic cytology specimens. Gut 2018;67:1942-9. [Crossref] [PubMed]
- Iyer PG, Taylor WR, Johnson ML, et al. Highly Discriminant Methylated DNA Markers for the Non-endoscopic Detection of Barrett's Esophagus. Am J Gastroenterol 2018;113:1156-66. [Crossref] [PubMed]
- Thrift AP. Barrett's Esophagus and Esophageal Adenocarcinoma: How Common Are They Really? Dig Dis Sci 2018;63:1988-96. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- El-Serag HB, Naik AD, Duan Z, et al. Surveillance endoscopy is associated with improved outcomes of oesophageal adenocarcinoma detected in patients with Barrett's oesophagus. Gut 2016;65:1252-60. [Crossref] [PubMed]
- Shaheen NJ, Falk GW, Iyer PG, et al. ACG Clinical Guideline: Diagnosis and Management of Barrett's Esophagus. Am J Gastroenterol 2016;111:30-50. [Crossref] [PubMed]
- Whiteman DC, Appleyard M, Bahin FF, et al. Australian clinical practice guidelines for the diagnosis and management of Barrett's esophagus and early esophageal adenocarcinoma. J Gastroenterol Hepatol 2015;30:804-20. [Crossref] [PubMed]
- Sami SS, Iyer PG, Pophali P, et al. Acceptability, Accuracy, and Safety of Disposable Transnasal Capsule Endoscopy for Barrett's Esophagus Screening. Clin Gastroenterol Hepatol 2019;17:638-646.e1. [Crossref] [PubMed]
- Jobe BA, Hunter JG, Chang EY, et al. Office-based unsedated small-caliber endoscopy is equivalent to conventional sedated endoscopy in screening and surveillance for Barrett's esophagus: a randomized and blinded comparison. Am J Gastroenterol 2006;101:2693-703. [Crossref] [PubMed]
- Bhardwaj A, Hollenbeak CS, Pooran N, et al. A meta-analysis of the diagnostic accuracy of esophageal capsule endoscopy for Barrett's esophagus in patients with gastroesophageal reflux disease. Am J Gastroenterol 2009;104:1533-9. [Crossref] [PubMed]
- Gora MJ, Sauk JS, Carruth RW, et al. Tethered capsule endomicroscopy enables less invasive imaging of gastrointestinal tract microstructure. Nat Med 2013;19:238-40. [Crossref] [PubMed]
- Ramirez FC, Akins R, Shaukat M. Screening of Barrett's esophagus with string-capsule endoscopy: a prospective blinded study of 100 consecutive patients using histology as the criterion standard. Gastrointest Endosc 2008;68:25-31. [Crossref] [PubMed]
- Vaughan TL, Fitzgerald RC. Precision prevention of oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2015;12:243-8. [Crossref] [PubMed]
- Spechler SJ, Katzka DA, Fitzgerald RC. New Screening Techniques in Barrett's Esophagus: Great Ideas or Great Practice? Gastroenterology 2018;154:1594-601. [Crossref] [PubMed]
- Ross-Innes CS, Chettouh H, Achilleos A, et al. Risk stratification of Barrett's oesophagus using a non-endoscopic sampling method coupled with a biomarker panel: a cohort study. Lancet Gastroenterol Hepatol 2017;2:23-31. [Crossref] [PubMed]
- Shah AK, Hartel G, Brown I, et al. Evaluation of Serum Glycoprotein Biomarker Candidates for Detection of Esophageal Adenocarcinoma and Surveillance of Barrett's Esophagus. Mol Cell Proteomics 2018;17:2324-34. [Crossref] [PubMed]
- Pavlov K, Kluiver J, Meijer C, et al. Circulating miRNAs in patients with Barrett's esophagus, high-grade dysplasia and esophageal adenocarcinoma. J Gastrointest Oncol 2018;9:1150-6. [Crossref] [PubMed]
- Chan DK, Zakko L, Visrodia KH, et al. Breath Testing for Barrett's Esophagus Using Exhaled Volatile Organic Compound Profiling With an Electronic Nose Device. Gastroenterology 2017;152:24-6. [Crossref] [PubMed]
- Smith MS, Cash B, Konda V, et al. Volumetric laser endomicroscopy and its application to Barrett’s esophagus: results from a 1,000 patient registry. Dis Esophagus 2019. [Epub ahead of print]. [Crossref] [PubMed]
Cite this article as: Shah AK, Joshi V, Hartel G, Barbour AP, Hill MM. To BE or not to BE: non-invasive screening for Barrett’s esophagus, dysplasia and adenocarcinoma. Transl Gastroenterol Hepatol 2019;4:31.


