Splicing alterations contributing to cancer hallmarks in the liver: central role of dedifferentiation and genome instability
Introduction
Cell fate and identity depend on the expression of a specific array of proteins at a precise moment. In eukaryotes the expression of multi-exon genes requires the efficient and correct removal or splicing of introns by the spliceosome, a highly flexible and reversible multiprotein enzyme. Alternative splicing affects 95% of genes and allows the generation, in a cell-type specific manner, of different mRNA isoforms from a single gene coding for proteins with even opposing functions (1). Alternative splicing can also modulate gene expression through for instance the inclusion or exclusion of poison exons able to activate non-sense mediated mRNA decay (NMD) (2-4). Therefore, transcription and alternative splicing are two tightly regulated processes responsible for the diversity of the proteome. The output of a splicing event depends on multiple factors including (I) cis-acting sequence motifs at splice sites at the exon-intron boundaries required for spliceosome assembly, and at the splicing enhancer and repressor motifs within the pre-mRNA which are recognized by RNA-binding proteins (RBP); (II) the concentration and availability of a large array of trans-acting regulatory splicing factors or RBPs able to bind the enhancer and repressor motifs and to modulate spliceosome activity and splice site selection; and (III) the kinetic competition between different spliceosome assembly pathways (5-8). Two key families of RBPs, the serine/arginine‐rich proteins (SR proteins) and the heterogeneous nuclear ribonucleoproteins (hnRNPs), regulate splicing in a synergic or competitive context-dependent manner and through many different mechanisms (5,7,9).
Alterations of splicing have been implicated in the development of different diseases including cancer (10). Malignant cell transformation requires the acquisition of several neoplastic capabilities through the activation of oncogenes and the inactivation of tumor suppressor genes (11). Differential splicing of specific genes and splicing factor alterations are present in most types of tumors and can be associated with each of the cancer hallmarks identified by Hanahan and Weinberg (6,9,12-14) In fact, the network of alternatively spliced transcripts is reprogrammed in cancer cells (13). These alterations can be associated with the presence of mutations in cis-acting splice sites or regulatory motifs and/or in the coding sequence of trans-acting splicing factors. However, and importantly, they can also be due to changes, even modest, in the expression, function and location of unmutated splicing factors (9,13) which can for instance be induced by the activation of specific signaling pathways (5,15).
In the last years it has been demonstrated that the splicing machinery, including spliceosome components and RBPs, regulates many relevant cellular processes in a splicing-independent manner. These processes include the maintenance of genome integrity by preventing the formation of RNA-DNA hybrids (R-loops) and by influencing the DNA damage response (DDR), transcription elongation and termination, mRNA nuclear export and translation-dependent non-sense mRNA decay response (NMD) (9,10). Importantly, these observations reveal the relevance that subtle changes in the abundance, post-translational modifications and/or subcellular localization of splicing factors and/or spliceosomal components may have, apart from splicing and gene expression, on very relevant cellular events which are central to the process of carcinogenesis.
Alternative splicing in HCC
Hepatocellular carcinoma (HCC), the most frequent tumor of the liver, develops in more than 80% of cases on a chronically damaged organ where hepatic functions have been lost. Molecularly, HCCs are very heterogeneous and different subclasses have been described according to the genetic alterations and biomarkers detected, in an attempt to improve the management of patients and the discovery of effective therapeutic strategies nowadays still elusive (16,17).
Alternative splicing is also emerging as a relevant player in the progression of liver disease (15,18) and recent high throughput studies have described the landscape of aberrant alternative splicing events in HCC (19-21). As described above, changes in splice variants may be associated with mutations on the splicing recognition motifs or with the dysregulation of splicing factors. Mutations, mis-localization and alterations in the level of expression of splicing factors have been described in HCC. For instance, the gene amplification and up-regulation of the spliceosome component splicing factor 3b subunit 4 (SF3B4) have been detected in precancerous lesions of HCC, being suggested as early-stage diagnostic markers and correlating with poor prognosis in HCC (22). A recent genome-wide study characterized the genetic alterations of RBPs in HCC and stablished the perturbations in the protein-RNA regulatory interactome in HCC (23). This study showed that somatic mutations are enriched in RBP binding sites and identified some interactions related to specific subtypes of HCC.
Regarding changes in localization, the cytosolic retention of the SR-protein SRSF3 through its interaction with the hepatitis B virus protein HBx, correlates with poor overall survival of HCC patients. Importantly, it has been associated with the aberrant splicing and up-regulation of an oncogenic truncated splice-isoform of CCDC50, an effector of epidermal growth factor (EGF)-mediated cell signaling implicated in the oncogenic progression of HCC (24).
Both up-regulation and down-regulation of splicing factors have been reported in HCC. hnRNPA1 and A2 (25) are upregulated in HCC and the induction of hnRNPC (20), hnRNPH1 and H2 (26), SRSF2 (27) and PTBP3 (28) has been correlated with poor prognosis. On the other hand, and as reviewed below, the downregulation of SRSF3 (29), SLU7 (30) and ESRP2 (31) has been also described in HCC.
In the next sections we will focus on selected altered splicing events. We will provide a new perspective of the changes in splicing factors in relation with the acquisition of specific cancer hallmarks during the process of hepatocarcinogenesis.
Alterations of splicing and hepatocyte de-differentiation in hepatocarcinogenesis
Epigenetic modifications, changes in the levels of expression of transcription factors, and regulation of mRNA processing are crucial for genome reprogramming during development and for the establishment of tissue-specific gene expression (32). Accordingly, the transition from the proliferative fetal liver to the metabolic functional postnatal organ and the fully differentiated adult liver is regulated not only by changes in transcription factors (33,34) but also by a relevant set of post-transcriptional splicing events (31). The importance of alternative splicing in the maintenance of the hepatic phenotype is evidenced by the fact that the liver, together with the brain and testis, is the organ with the greatest diversity in transcripts associated with alternative exon or splice site usage (35).
The liver is a highly specialized and differentiated organ, however as mentioned above HCC fundamentally develops on cirrhotic tissues where the characteristic hepatic functions have been significantly blunted. This progressive loss of functions is linked to changes in the profile of gene expression and switches in isozymes expression towards a more fetal-like and de-differentiated landscape. These changes are largely associated with alterations in the expression of transcription factors which include the inhibition of HNF4 and the induction of Wilms’ tumor 1 in human cirrhosis and HCC (36-38). In fact, enforced expression of HNF4α attenuates hepatic fibrosis (39), reverses terminal chronic hepatic failure (40), and blocks HCC occurrence in rats (41), while its depletion fosters hepatocarcinogenesis (42,43). Other example would be the dysregulation of the HIPPO/YAP cascade. Nuclear staining of the transcriptional co-activator YAP is detected in 50% of human HCCs suggesting a role for YAP activation (44). Accordingly, the activation of endogenous YAP perturbs hepatocyte differentiation and maintains this immature state in advanced tumors in mice (45). Interestingly, YAP silencing in mouse HCC restores hepatocyte differentiation and leads to tumor regression (45). As discussed below downregulation of the zinc finger transcription factor Krüppel-like factor 6 (KLF6), and its altered splicing pattern, is observed in early human hepatocarcinogenesis and is also related with hepatocellular de-differentiation (46).
More recently, it has been demonstrated that the preservation of a liver-specific transcriptional profile depends to a great extent on the correct expression of three splicing factors, the SR-protein SRSF3 (29), the pre-mRNA splicing factor SLU7 (30) and epithelial splicing regulatory protein 2 (ESRP2) (31). These studies show that the hepatic depletion or reduction of expression of SRSF3, SLU7 or ESRP2 in mice impact significantly on the mature and metabolically functional phenotype of the liver, changing not only the alternatively spliced transcriptome profile but also the rate of transcription of oncofetal genes such as alfa-fetoprotein (AFP) and the non-coding RNA H19, as well as metabolic and proliferation-related genes (29,30). In addition, the data demonstrates a complex cross-regulation among the different effectors and pathways. For instance, reduced expression of SLU7 results in altered splicing and diminished expression of SRSF3 and changes in the use of HNF4 promoter, from the adult-specific P1 promoter to the fetal/oncogenic P2 promoter (30,38). Importantly, in support of the relevance and pathological implications of these findings, the expression of SRSF3, SLU7 and ESRP2 is significantly reduced in human HCC (19,47,48), and that of SLU7 is also impaired in the preneoplastic cirrhotic liver (47). More specifically, it has been demonstrated that the hepatocyte-specific knockdown of SRSF3 in mice results in the spontaneous development of HCC (48), further emphasizing the relevance of hepatic de-differentiation in the process of hepatocarcinogenesis.
Recently, muscleblind-like-3 (MBNL3) has been identified as a liver oncofetal splicing factor expressed at high levels in fetal livers, silenced in adult livers and reexpressed in HCC tissues (49). Its oncogenic function has been linked to the inclusion of exon 4 into the lncRNA-PXN-AS1 and the subsequent upregulation of the cytoskeletal oncoprotein paxillin (PXN) (49).
The cell fate determinant NUMB is a direct transcriptional target of the WNT pathway and a negative regulator of NOTCH signaling promoting hepatocyte differentiation (50). The upregulation of an aberrant alternatively spliced isoform of NUMB after the inclusion of exon 12 (PRRL isoform) has been detected in HCC and it is associated with early recurrence and reduced overall survival after surgery (51). Mechanistically, this splicing event is inhibited by the splicing factor RBFOX2 and promoted by the cytoplasmic retention of the SR-protein kinase SRPK2 through its interaction with the chaperon HSP90 (51). In fact, the presence of NUMB PRRL isoform was proposed as a biomarker in HCC to stratify patients to be treated with HSP90-targeted drugs (51,52). In this same line, a differentiation therapy for HCC has been proposed using miR-148a mimics, which mediate hepatocyte differentiation through the upregulation of NUMB expression (53).
Many metabolism-related enzymes are expressed as cell-type specific isoforms regulated by alternative splicing. In fact, many de-differentiation events observed during the process of hepatocarcinogenesis represent switches of alternatively spliced metabolic enzyme isoforms. In general, mature liver-specific isoforms are replaced by fetal isoforms or isoforms normally expressed in other tissues (20,21,54).
Alterations of splicing and HCC metabolic reprogramming
As mentioned above, many metabolism-related genes are regulated by alternative splicing. Cancer is recognized as a disease of energetic metabolism and the metabolic reprogramming of cancer cells, including the use of glucose, glutamine and fructose is facilitated by changes in the expression of mutually exclusive alternatively spliced isoforms of metabolic enzymes (54).
Fructose is mainly catabolized in the liver by the high affinity enzyme fructokinase C (KHK-C) an isoform mainly expressed in hepatocytes generated by the incorporation of alternative exon 3C into the mRNA (55). However, in HCC cells RBP hnRNPH1/2 promotes the exclusion of exon 3C and the inclusion of exon 3A to generate the isoform KHK-A. This results in a reduced fructose metabolism rate preventing enhanced ROS generation and uncontrolled lipid production, along with promoting glucose-derived de novo nucleic acid synthesis through phosphorylation of phosphoribosyl pyrophosphate synthetase 1 (PRPS1) (54). Therefore, this splicing switch supports tumorigenesis and allows the coordinated regulation of glucose and fructose metabolism in HCC cells (54). Importantly, hnRNAPH1/2 expression is regulated by cMYC and the expression of cMYC, hnRNPH1/2 and KHK-A is correlated and significantly up-regulated in HCC tissues, being independent prognostic factors for overall survival (54).
Aerobic glycolysis or Warburg effect plays a crucial role in the process of carcinogenesis (11). Pyruvate kinase (PK) catalyzes the last committed step in glycolysis, the conversion of phosphoenolpyruvate (PEP) to pyruvate. The main isozyme expressed in the liver is PKL, however, alternative splicing of the isozyme pyruvate kinase M (PKM) is an important determinant of the Warburg effect of cancer cells versus differentiated cells (56). Two isoforms PKM1 and PKM2 are expressed through mutually exclusive alternative splicing of exons 9 and 10 (57), being PKM2 expressed during embryogenesis, tissue regeneration, and tumor development (58). Interestingly, PKM2 overexpression in HCC is associated with poor prognosis (59). Mechanistically, hnRNPA1/hnRNPA2 have been shown to inhibit exon 9 inclusion, favoring PKM2 expression (60). As mentioned before hnRNPA1/hnRNPA2 expression is induced in HCC (25) which could be due to cMYC activation (60) or SLU7 downregulation (30) both events observed in HCC (47,54).
The alternative splicing of exon 11 into the insulin receptor (IR) mRNA is developmentally regulated in a tissue-specific manner. The fetal liver expresses IR-A isoform and the skipping of exon 11 confers a higher affinity not only for insulin but also for insulin like growth factor II (IGF-II), a growth factor mainly implicated in proliferation (61). The differentiated adult liver expresses almost exclusively the IR-B isoform containing exon 11 and, being involved in the metabolic effects of insulin (61). Interestingly, the ratio IR-A/IR-B is significantly increased in HCC and in a model of hepatocarcinogenesis in rats (25) suggesting a role in the transformation of hepatocytes. Mechanistically this dysregulation of IR splicing can be induced by the upregulation of hnRNPA1 (25) or the downregulation of SRSF3 (29) or SLU7 (30), all three events observed in human HCC (25,29,47).
Alterations of splicing and genome instability in HCC
Genomic instability is considered an enabling characteristic of cancer cells endowing them with genetic alterations that support their growth (11). Genome instability can be the result of increased DNA damage and accumulation of mutations or mitotic errors associated with chromosome alterations. Alternative splicing has been implicated in the regulation of both processes and splicing factors are emerging as gatekeepers of genome stability (62,63).
Chromosome instability (CIN) affecting the number and structure of chromosomes is one of the most common alterations in HCC (64). Several splicing events affecting proteins implicated in the mitotic spindle checkpoint (MSC), which is responsible for inducing mitosis arrest to prevent chromosome mis-segregation, have been described in HCC. For instance, an aberrant isoform deleted in exon 4 (MAD1beta) of the mitotic arrest deficient 1 (MAD1) gene is induced in 24% of HCCs (65). MAD1beta sequesters MAD2 in the cytoplasm preventing its function as controller in the mitotic checkpoint and inducing the formation of chromosome bridges and aneuploidy (65). Its relevance in hepatocarcinogenesis is supported by the fact that heterozygous deletion of MAD1 in mice results in the development of HCC (66). The splicing factors implicated in this aberrant event have not been characterized yet.
Another important player in MSC is the serine/threonine kinase Aurora B (AURKB) which is overexpressed in HCC (67). Importantly, aberrant splicing isoforms are also induced, and in particular a small percentage of patients overexpress a variant lacking exon 6 (AURKB-Sv2) which correlates with poor prognosis (67). This isoform, deprived of the kinase activity, could act as dominant negative of AURKB and participate in the induction of CIN (67). Again, the mechanisms and splicing factors implicated in this aberrant event have not been elucidated.
Correct sister chromatid cohesion (SCC) is essential to secure the proper segregation of chromosomes during mitosis. Shugoshins (SGO) are proteins required for the correct cohesion of centromeres (68) and the heterozygous deletion of SGO1 in mice induces CIN and the development of HCC (69). Importantly a splicing isoform (sSGO1) lacking exon 6 is located at the centrosomes instead of at the centromeres being a guardian of centriole cohesion (70), and sSGO1 overexpression induces multipolar cells and chromosome mis-segregation (71). The induction of SGO1 expression has been described in HCC (72), however the characterization of the splicing isoforms expressed and in particular sSGO1 expression has not been addressed. Our observations demonstrate that SLU7 knockdown induces the depletion of exon 6 and the induction of sSGO1 (73), suggesting that sSGO1 expression deserves further studies in HCC and could be implicated in the induction of CIN. Our data also shows that SLU7 plays a more general role in maintaining SCC and securing chromosome stability. Sororin (CDCA5) is essential to maintain SCC (74). We have demonstrated that SLU7 downregulation induces the aberrant incorporation of introns 1 and 2 into the mRNA of sororin, inducing its degradation by the NMD machinery and resulting in reduced protein levels (73). Consequently, SLU7 silencing results in defects in SCC and mitosis arrest in HCC cells (73).
DNA damage occurs mainly because of errors during replication, exposure to oxidative stress and damaging agents, and RNA transcription-dependent formation of RNA-DNA hybrids or R-loops (63,75). DNA damage is present in the liver of cirrhotic patients and in HCC, as evidenced by the increased detection of the biomarker γH2AX (76). DNA damage has been demonstrated as determinant in the induction of HCC in mice (77,78). Cells are endowed with systems to sense and respond to DNA damage, and a potent DNA damage repair (DDR) machinery is activated in the cell to prevent the fixation of mutations. Recently the existence of an important interplay between the DNA damage response and RNA processing has been recognized (63). RBPs play an important role regulating not only the transcription and splicing of DDR sensors and effectors, but also controlling directly the DDR in a splicing-independent manner (63,79,80). RNA-transcription is also a source of genome instability through the formation of RNA-DNA hybrids (R-loops). These R-loops are formed between the nascent mRNA and the template strand of the DNA, leaving the coding DNA strand exposed to damaging agents (81). R-loops are generated physiologically and are processed by RNase H1, however they can accumulate under certain conditions including diminished expression of RBPs, promoting mutations, recombination and chromosome rearrangements (63,81,82). Recently it has been shown that RNAse H1 depletion in the liver of mice results in R-loops accumulation and the impairment of liver function (83). The depletion of SRSF1 and SRSF3 has been associated with the accumulation of R-loops (84). The downregulation or mislocalization of SRSF3 observed in HCC (24,48) could therefore participate in the induction of R-loops promoting genome instability.
Alterations of splicing and cell cycle progression in HCC
As mentioned above, RNA processing and alternative splicing affect most of the genes expressed in humans (1). The splicing landscape is reprogrammed in cancer cells (13) and alterations of splicing represent one of the mechanisms used by cancer cells to activate oncogenes or inactivate tumor suppressor genes (TSGs). In fact, global profiling of alternative RNA splicing events in HCC reveals the existence of alternative splicing signatures associated with different types of HCC (20). As already discussed, these changes can be linked to mutations in specific splicing regulation sites or to changes in RNA splicing factors that act as oncoproteins or TSGs (9,15). In fact, many of the already discussed alterations of splicing observed in HCC such as CCDC50, PXN, IR-A and PKM2, affect cell cycle progression inducing cell proliferation and/or cell survival. In this section we will describe two examples of well-characterized pathways connecting altered cell signaling with dysregulated splicing factors expression, and the inactivation of TSG transcription factors in HCC.
The transcription factor and tumor suppressor gene KLF6 regulates cell differentiation, proliferation and survival, and it is expressed as four splicing isoforms (85). KLF6-SV1 isoform lacks the three zinc finger DNA binding domains acting as dominant-negative and antagonizing KLF6, leading for instance to decreased p21 expression and increased cell growth (86). Reduced KLF6 expression has been described in HCC and an increased SV1/KLF6 ratio is associated with aggressive clinical behavior both in human and in mice (46,87). The mechanisms implicated in the splicing inactivation of KLF6 have been described. SRSF1 is required to express the full length KLF6 mRNA (88), and SV1 induction is associated with RAS/PI3K/AKT activation (88) and hepatocyte growth factor signaling (HGF) (89) in HCC cells. Mechanistically, HGF induces c-MET and PI3K/AKT signaling and downregulates SRSF3 expression which is required for the correct splicing and expression of SRSF1 (89). SRSF1 mRNA degradation by NMD and its protein reduction favor KLF6 splicing, SV1 isoform expression and its oncogenic properties (89). All together these data suggest that c-MET activation, SRSF3 downregulation, and KLF6-SV1 induction could represent coordinated events useful to identify subgroups of HCC patients with specific targetable alterations.
The tumor suppressor gene P73 belongs to the P53 gene family of transcription factors (90). A large number of isoforms are generated through alternative promoter usage and multiple alternative splicing events. Isoforms lacking the transactivation domain (TA) behave as dominant negative inhibitors of both TSGs, P73 and P53, displaying oncogenic properties (91). Interestingly, transgenic mice expressing an isoform lacking exons 2 and 3 in hepatocytes spontaneously develop HCC (92). We have demonstrated that the growth factor amphiregulin (AREG) activates EGFR and JNK1 to downregulate the expression of the splicing factor SLU7, which is responsible for the correct incorporation of exon 2 into P73 mRNA (47). The relevance of these results is supported by the fact that AREG expression is induced in parallel to the downregulation of SLU7 expression and the induction of the p73 isoform defective in exon 2 (Ex2p73) not only in HCC but also in the preneoplastic cirrhotic liver (47).
Conclusions and perspectives
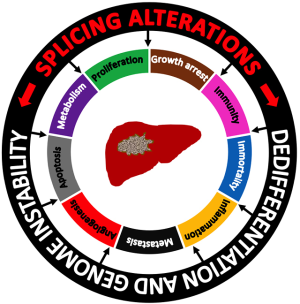
HCCs are molecularly heterogeneous tumors, and this complexity is to a great extent responsible for their poor response to conventional and targeted therapies (16,93). The information summarized in this review indicates that imbalanced expression of splicing factors can be a relevant source for this heterogeneity. Moreover, we have also illustrated how these alterations may play a driver role in hepatocarcinogenesis by impinging on the general hallmarks of cancer. Having in mind the natural history of HCC we focused on two pathogenic features that are characteristic of liver tumors: chromosomal instability and phenotypic dedifferentiation. We highlight mechanisms connecting splicing derangement with these two processes and the enabling capacities acquired by liver cells along their neoplastic transformation (Figure 1). A thorough understanding of the alterations in the splicing machinery may also help to design novel therapeutic strategies. Indeed, relevant progress has been made in the identification of small molecules that can interfere with the activity of splicing factors at different levels, from their expression to their enzymatic or structural activities (14). However, their precise mechanisms of action are not always well characterized, and although cancer cells seem more susceptible to the inhibition of the splicing machinery than normal cells (94) unexpected toxicities may occur (95). RNA-based therapeutics using splice-switching antisense oligonucleotides (ASO) are also actively pursued (14). These ASO can target specific components of the splicing machinery, potentially avoiding toxic effects. However, their efficient delivery to target tissues is still a challenge (96), and in the case of HCC a critical one given the profound histological alterations of the cirrhotic liver on which HCCs develop. From a translational point of view, and as discussed in previous sections, the identification of splicing isoforms specific of HCC cells may provide robust biomarkers of the disease (7). Moreover, these HCC-specific variants could constitute tumor associated antigens that may be harnessed for the development of cancer vaccines and immunotherapy strategies against liver tumors (97).
Acknowledgements
Work in the authors’ laboratory is supported by CIBERehd, Instituto de Salud Carlos III; Grant MINECO SAF2016-75972-R; HEPACARE Project from Fundación La Caixa; ADA fellowship to M Jimenez; AECC post-doctoral fellowship to M Arechederra; Fundación Eugenio Rodríguez Pascual; Fundación Echebano; Fundación Mario Losantos; Fundación M Torres and Mr. Eduardo Ávila and Mr. Sergio Durá contributions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008;40:1413-5. [Crossref] [PubMed]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 2002;3:285-98. [Crossref] [PubMed]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci 2008;33:385-93. [Crossref] [PubMed]
- da Costa PJ, Menezes J, Romão L. The role of alternative splicing coupled to nonsense-mediated mRNA decay in human disease. Int J Biochem Cell Biol 2017;91:168-75. [Crossref] [PubMed]
- Fu XD, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 2014;15:689-701. [Crossref] [PubMed]
- Anczuków O, Krainer AR. Splicing-factor alterations in cancers. RNA 2016;22:1285-301. [Crossref] [PubMed]
- Wan Y, Larson DR. Splicing heterogeneity: separating signal from noise. Genome Biol 2018;19:86. [Crossref] [PubMed]
- Dvinge H. Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett 2018;592:2987-3006. [Crossref] [PubMed]
- Dvinge H, Kim E, Abdel-Wahab O, et al. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 2016;16:413-30. [Crossref] [PubMed]
- Carey KT, Wickramasinghe VO. Regulatory Potential of the RNA Processing Machinery: Implications for Human Disease. Trends Genet 2018;34:279-90. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Singh B, Eyras E. The role of alternative splicing in cancer. Transcription 2017;8:91-8. [Crossref] [PubMed]
- El Marabti E, Younis I. The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front Mol Biosci 2018;5:80. [Crossref] [PubMed]
- Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 2018;9. [Crossref] [PubMed]
- Berasain C, Elizalde M, Urtasun R, et al. Alterations in the expression and activity of pre-mRNA splicing factors in hepatocarcinogenesis. Hepat Oncol 2014;1:241-52. [Crossref] [PubMed]
- Zucman-Rossi J, Villanueva A, Nault JC, et al. Genetic Landscape and Biomarkers of Hepatocellular Carcinoma. Gastroenterology 2015;149:1226-1239.e4. [Crossref] [PubMed]
- Llovet JM, Montal R, Sia D, et al. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol 2018;15:599-616. [Crossref] [PubMed]
- Webster NJG. Alternative RNA Splicing in the Pathogenesis of Liver Disease. Front Endocrinol (Lausanne) 2017;8:133. [Crossref] [PubMed]
- Zhang L, Liu X, Zhang X, et al. Identification of important long non-coding RNAs and highly recurrent aberrant alternative splicing events in hepatocellular carcinoma through integrative analysis of multiple RNA-Seq datasets. Mol Genet Genomics 2016;291:1035-51. [Crossref] [PubMed]
- Tremblay MP, Armero VES, Allaire A, et al. Global profiling of alternative RNA splicing events provides insights into molecular differences between various types of hepatocellular carcinoma. BMC Genomics 2016;17:683. [Crossref] [PubMed]
- Li S, Hu Z, Zhao Y, et al. Transcriptome-Wide Analysis Reveals the Landscape of Aberrant Alternative Splicing Events in Liver Cancer. Hepatology 2018. [Epub ahead of print]. [PubMed]
- Shen Q, Nam SW. SF3B4 as an early-stage diagnostic marker and driver of hepatocellular carcinoma. BMB Rep 2018;51:57-8. [Crossref] [PubMed]
- Li Y, McGrail DJ, Xu J, et al. MERIT: Systematic analysis and characterization of Mutational Effect on RNA Interactome Topology. Hepatology 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Hong W, Zhang CZ, Lu SX, et al. A CCDC50 splice variant is modulated by SRSF3 and promotes hepatocellular carcinoma via the Ras signaling pathway. Hepatology 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Chettouh H, Fartoux L, Aoudjehane L, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res 2013;73:3974-86. [Crossref] [PubMed]
- Li X, Qian X, Peng LX, et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat Cell Biol 2016;18:561-71. [Crossref] [PubMed]
- Luo C, Cheng Y, Liu Y, et al. SRSF2 Regulates Alternative Splicing to Drive Hepatocellular Carcinoma Development. Cancer Res 2017;77:1168-78. [Crossref] [PubMed]
- Yang X, Qu S, Wang L, et al. PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612. Oncogene 2018;30:444. [PubMed]
- Sen S, Jumaa H, Webster NJG. Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat Commun 2013;4:1336. [Crossref] [PubMed]
- Elizalde M, Urtasun R, Azkona M, et al. Splicing regulator SLU7 is essential for maintaining liver homeostasis. J Clin Invest 2014;124:2909-20. [Crossref] [PubMed]
- Bhate A, Parker DJ, Bebee TW, et al. ESRP2 controls an adult splicing programme in hepatocytes to support postnatal liver maturation. Nat Commun 2015;6:8768. [Crossref] [PubMed]
- Fiszbein A, Kornblihtt AR. Alternative splicing switches: Important players in cell differentiation. Bioessays 2017;39. [Crossref] [PubMed]
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell 2010;18:175-89. [Crossref] [PubMed]
- Lemaigre FP. Mechanisms of liver development: concepts for understanding liver disorders and design of novel therapies. Gastroenterology 2009;137:62-79. [Crossref] [PubMed]
- Yeo G, Holste D, Kreiman G, et al. Variation in alternative splicing across human tissues. Genome Biol 2004;5:R74. [Crossref] [PubMed]
- Avila MA, Berasain C, Torres L, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol 2000;33:907-14. [Crossref] [PubMed]
- Berasain C, Herrero JI, García-Trevijano ER, et al. Expression of Wilms' tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology 2003;38:148-57. [Crossref] [PubMed]
- Berasain C, Avila MA. Regulation of hepatocyte identity and quiescence. Cell. Mol. Life Sci 2015;72:3831-51. [Crossref] [PubMed]
- Yue HY, Yin C, Hou JL, et al. Hepatocyte nuclear factor 4alpha attenuates hepatic fibrosis in rats. Gut 2010;59:236-46. [Crossref] [PubMed]
- Nishikawa T, Bell A, Brooks JM, et al. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest 2015;125:1533-44. [Crossref] [PubMed]
- Ning B-F, Ding J, Yin C, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res 2010;70:7640-51. [Crossref] [PubMed]
- Lazarevich NL, Cheremnova OA, Varga EV, et al. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology 2004;39:1038-47. [Crossref] [PubMed]
- Walesky C, Edwards G, Borude P, et al. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology 2013;57:2480-90. [Crossref] [PubMed]
- Tschaharganeh DF, Chen X, Latzko P, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology 2013;144:1530-1542.e12. [Crossref] [PubMed]
- Fitamant J, Kottakis F, Benhamouche S, et al. YAP Inhibition Restores Hepatocyte Differentiation in Advanced HCC, Leading to Tumor Regression. Cell Rep 2015;10:1692-707. [Crossref] [PubMed]
- Kremer-Tal S, Narla G, Chen Y, et al. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol 2007;46:645-54. [Crossref] [PubMed]
- Castillo J, Goñi S, Latasa MU, et al. Amphiregulin induces the alternative splicing of p73 into its oncogenic isoform DeltaEx2p73 in human hepatocellular tumors. Gastroenterology 2009;137:1805-15.e1-4.
- Sen S, Langiewicz M, Jumaa H, et al. Deletion of serine/arginine‐rich splicing factor 3 in hepatocytes predisposes to hepatocellular carcinoma in mice. Hepatology 2015;61:171-83. [Crossref] [PubMed]
- Yuan JH, Liu XN, Wang TT, et al. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol 2017;19:820-32. [Crossref] [PubMed]
- Boulter L, Govaere O, Bird TG, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18:572-9. [Crossref] [PubMed]
- Lu Y, Xu W, Ji J, et al. Alternative splicing of the cell fate determinant Numb in hepatocellular carcinoma. Hepatology 2015;62:1122-31. [Crossref] [PubMed]
- Zhao J, Gray SG, Wabitsch M, et al. The therapeutic properties of resminostat for hepatocellular carcinoma. Oncoscience 2018;5:196-208. [Crossref] [PubMed]
- Jung KH, Zhang J, Zhou C, et al. Differentiation therapy for hepatocellular carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology 2016;63:864-79. [Crossref] [PubMed]
- Kozlovski I, Siegfried Z, Amar-Schwartz A, et al. The role of RNA alternative splicing in regulating cancer metabolism. Hum Genet 2017;136:1113-27. [Crossref] [PubMed]
- Hayward BE, Bonthron DT. Structure and alternative splicing of the ketohexokinase gene. Eur J Biochem 1998;257:85-91. [Crossref] [PubMed]
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev 2010;24:2343-64. [Crossref] [PubMed]
- Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem 1986;261:13807-12. [PubMed]
- Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol 2011;43:969-80. [Crossref] [PubMed]
- Liu Y, Wu H, Mei Y, et al. Clinicopathological and prognostic significance of PKM2 protein expression in cirrhotic hepatocellular carcinoma and non-cirrhotic hepatocellular carcinoma. Sci Rep 2017;7:15294. [Crossref] [PubMed]
- David CJ, Chen M, Assanah M, et al. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010;463:364-8. [Crossref] [PubMed]
- Belfiore A, Frasca F, Pandini G, et al. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr Rev 2009;30:586-623. [Crossref] [PubMed]
- López-Saavedra A, Herrera LA. The role of alternative mRNA splicing in chromosome instability. Mutat Res 2010;705:246-51. [Crossref] [PubMed]
- Naro C, Bielli P, Pagliarini V, et al. The interplay between DNA damage response and RNA processing: the unexpected role of splicing factors as gatekeepers of genome stability. Front Genet. 2015;6:142. [Crossref] [PubMed]
- Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol 2016;22:9069-95. [Crossref] [PubMed]
- Sze KM-F, Ching Y-P, Jin D-Y, et al. Role of a novel splice variant of mitotic arrest deficient 1 (MAD1), MAD1beta, in mitotic checkpoint control in liver cancer. Cancer Res 2008;68:9194-201. [Crossref] [PubMed]
- Iwanaga Y, Chi YH, Miyazato A, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res 2007;67:160-6. [Crossref] [PubMed]
- Yasen M, Mizushima H, Mogushi K, et al. Expression of Aurora B and alternative variant forms in hepatocellular carcinoma and adjacent tissue. Cancer Sci 2009;100:472-80. [Crossref] [PubMed]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 2004;118:567-78. [Crossref] [PubMed]
- Yamada HY, Kumar G, Zhang Y, et al. Systemic chromosome instability in Shugoshin-1 mice resulted in compromised glutathione pathway, activation of Wnt signaling and defects in immune system in the lung. Oncogenesis 2016;5. [Crossref] [PubMed]
- Wang X, Yang Y, Duan Q, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell 2008;14:331-41. [Crossref] [PubMed]
- Suzuki H, Akiyama N, Tsuji M, et al. Human Shugoshin mediates kinetochore-driven formation of kinetochore microtubules. Cell Cycle 2006;5:1094-101. [Crossref] [PubMed]
- Wang LH, Yen CJ, Li TN, et al. Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget 2015;6:2023-33. [PubMed]
- Jimenez M, Urtasun R, Elizalde M, et al. SLU7 is a survival factor for cancer cells working as a mitotic regulator. J Hepatol 2017;66:S645-654. [Crossref]
- Zhang N, Pati D. Sororin is a master regulator of sister chromatid cohesion and separation. Cell Cycle 2012;11:2073-83. [Crossref] [PubMed]
- Mikolaskova B, Jurcik M, Cipakova I, et al. Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways. Curr Genet 2018;64:971-83. [Crossref] [PubMed]
- Matsuda Y, Wakai T, Kubota M, et al. DNA damage sensor γ -H2AX is increased in preneoplastic lesions of hepatocellular carcinoma. ScientificWorldJournal 2013;2013. [Crossref] [PubMed]
- Barash H, R, Gross E, Edrei Y, et al. Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc Natl Acad Sci U S A 2010;107:2207-12. [Crossref] [PubMed]
- Boege Y, Malehmir M, Healy ME, et al. A Dual Role of Caspase-8 in Triggering and Sensing Proliferation-Associated DNA Damage, a Key Determinant of Liver Cancer Development. Cancer Cell 2017;32:342-359.e10. [Crossref] [PubMed]
- Montecucco A, Biamonti G. Pre-mRNA processing factors meet the DNA damage response. Front Genet 2013;4:102. [Crossref] [PubMed]
- Wickramasinghe VO, Venkitaraman AR. RNA Processing and Genome Stability: Cause and Consequence. Molecular Cell 2016;61:496-505. [Crossref] [PubMed]
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Molecular Cell 2012;46:115-24. [Crossref] [PubMed]
- Kim N, Jinks-Robertson S. Transcription as a source of genome instability. Nat Rev Genet 2012;13:204-14. [Crossref] [PubMed]
- Lima WF, Murray HM, Damle SS, et al. Viable RNaseH1 knockout mice show RNaseH1 is essential for R loop processing, mitochondrial and liver function. Nucleic Acids Res 2016;44:5299-312. [Crossref] [PubMed]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005;122:365-78. [Crossref] [PubMed]
- DiFeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat 2009;12:1-7. [Crossref] [PubMed]
- Narla G, DiFeo A, Reeves HL, et al. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res 2005;65:1213-22. [Crossref] [PubMed]
- Vetter D, Cohen-Naftaly M, Villanueva A, et al. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology 2012;56:1361-70. [Crossref] [PubMed]
- Yea S, Narla G, Zhao X, et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology 2008;134:1521-31. [Crossref] [PubMed]
- Muñoz Ú, Puche JE, Hannivoort R, et al. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol Cancer Res 2012;10:1216-27. [Crossref] [PubMed]
- Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 2007;7:165-8. [Crossref] [PubMed]
- Oswald C, Stiewe T. In good times and bad: p73 in cancer. Cell Cycle 2008;7:1726-31. [Crossref] [PubMed]
- Tannapfel A, John K, Mise N, et al. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis 2008;29:211-8. [Crossref] [PubMed]
- Gerbes A, Zoulim F, Tilg H, et al. Gut roundtable meeting paper: selected recent advances in hepatocellular carcinoma. Gut 2018;67:380-8. [Crossref] [PubMed]
- Urtasun R, Elizalde M, Azkona M, et al. Splicing regulator SLU7 preserves survival of hepatocellular carcinoma cells and other solid tumors via oncogenic miR-17-92 cluster expression. Oncogene 2016;35:4719-29. [Crossref] [PubMed]
- Agrawal AA, Yu L, Smith PG, et al. Targeting splicing abnormalities in cancer. Curr Opin Genet Dev 2018;48:67-74. [Crossref] [PubMed]
- Juliano RL. The delivery of therapeutic oligonucleotides. Nucleic Acids Res 2016;44:6518-48. [Crossref] [PubMed]
- Obeid JM, Kunk PR, Zaydfudim VM, et al. Immunotherapy for hepatocellular carcinoma patients: is it ready for prime time? Cancer Immunol Immunother 2018;67:161-74. [Crossref] [PubMed]
Cite this article as: Jimenez M, Arechederra M, Ávila MA, Berasain C. Splicing alterations contributing to cancer hallmarks in the liver: central role of dedifferentiation and genome instability. Transl Gastroenterol Hepatol 2018;3:84.


