Targeting autophagy in liver cancer
Introduction
Since the discovery of autophagy mechanism in 1956 (1), several studies have further identified autophagy as an intriguing mechanism responsible for catabolizing all the subcellular organelles and particles that could be recycled by the cell for its survival. Thus, autophagy vacuoles need the fusion with the lysosome to execute the degradation process (2).
Alteration and/or impairment of autophagy process have been shown to be implicated in the development of neurodegenerative diseases of central nervous system (3,4) and cancer (5,6).
Additionally, autophagy has been implicated in liver steatosis, especially nonalcoholic steatohepatitis (NASH), and exerts a key role during hepatitis B virus (HBV) and hepatitis C virus (HCV) hepatocytes infection. Furthermore, the alteration of autophagy process is responsible for the development of solid malignancies, e.g., breast cancer and liver cancer.
Interestingly, several key players of autophagy have been implicated in liver tumorigenesis. BECN1 (ATG6, Beclin-1) represents, for example, a haploinsufficient tumor suppressor; its heterozygous deletion causes the development of liver tumors in mice (6). ATG5, ATG7 and p62 (SQSTM1; sequestosome 1) have shown to exert tumorigenic role and their suppression reduces liver tumorigenesis (7,8).
In the last years, several studies have shown that some compounds are able to target autophagy and modulate the function of this metabolic process thus leading to block of cell proliferation and cell death in solid malignancies, e.g., liver cancer. For example, valproic acid and panobinostat, pan-deacetylase inhibitors (9,10), and the inhibitors of Cdc2-like kinase 1 (CLK1) (11) have shown the ability to induce autophagy. Still controversial is the role of sorafenib, a well-known tyrosine kinase inhibitor approved for the treatment of liver malignancies and other solid tumors, as modulator of autophagy in hepatocellular carcinoma (HCC) (12,13). Furthermore, unpublished data obtained from our group have shown the involvement of CUX1 (CUTL1, homeobox protein cut-like 1) in the autophagy process in liver cancer.
Recently, the discovery of PD-1/PD-L1 as druggable target in solid tumor with aggressive and metastatic potential has shown of being able to modulate autophagy (14). The potential of targeting autophagy via PD-1 in liver cancer could be a future promising therapy for this severe malignancy that need to be further investigated.
Autophagy discovery
The investigation of autophagy was determined by the initial discovery of the lysosome in 1950s (1).
At that time, Novikoff et al. firstly observed in 1956, by electron microscopy, the presence of lysosome-rich fractions in rat liver, which they called “dense bodies”. Also, they observed that these phosphatase acid-positive double membrane structures contain mitochondria and parts of the ER. But, they couldn’t verify the correlation between dense bodies and lysosomes (15).
In 1960 Essner and Novikoff proved that the dense bodies of human hepatic cells, so called hepatocellular pigments, are lysosome vesicles (16).
Ashford and Porter firstly described in rat liver cells the engulfment and lysosome-mediated degradation of various cytoplasmic components after treatment with glucagon. In addition, they hypothesized that lysosomes were such a kind of portions of cytoplasm including other organelles (17). Later de Duve introduced the term autophagosome to describe a double membrane organelle containing cytoplasmic components and the term Autophagy (“self-eating”, Greek) to describe the lysosomal digestion of own-cytoplasmic cellular components (18,19).
Arstila and Trump (2) firstly described that the autophagosome originate from the cisternae of ER. It represents an acid hydrolase-free double membrane vacuole. The transfer of hydrolytic enzymes into the autophagosome requires fusion of the autophagosome and lysosome. The new formed autolysosome enables the degradation of cytoplasmic elements by lysosomal enzymes and transforms from a double membrane to a single membrane structure depending on the maturation stage (2). Thereby, they characterized in principle the morphological development of autophagic flux. Despite of the catalytic role exerted by glucagon and starvation during autophagy, Pfeifer and Strauss demonstrated that food intake and insulin inhibit autophagy; thus, highlighting the link between autophagic activity and metabolism (20-22). For a long time, autophagy research was only based on morphological studies. No characteristic genes and proteins of autophagic machinery have been identified until Ohsumi discovered extensive autophagy induction through starvation in yeast cells (Saccharomyces cerevisiae) and started to use yeast as a model to study the molecular mechanisms and genes involved in autophagy (23,24). Up to now, 30 autophagy related (atg/ATG) genes of yeast (atg) and their mammalian homologs (ATG) were discovered (25). Interestingly, in 1999 a connection between autophagic activity and tumorigenesis was observed (26).
Autophagy in mammalians and humans
Cellular stocks of organelles, proteins and other sub-cellular components are continuously synthesized and degraded. One way to enable energy dependent processes such as protein synthesis is the intake of nutrients from the external environment through endocytosis (27). During nutrient starvation, cells rely on autophagy to provide energy and sustain cell homeostasis through degradation and recycling of cytosolic compartments (28).
Autophagy is differentiated into three types (I) chaperone-mediated autophagy, (II) micro-autophagy and (III) macro-autophagy, which all lead to lysosomal degradation of cytosolic components. Chaperone-mediated autophagy enables the selective recognition of cytosolic proteins by a chaperone and their delivery to the lysosomal membrane, where the proteins are unfolded and translocated into the lysosome (29). Micro-autophagy describes the direct engulfment of cytosolic proteins and organelles by the lysosome (29).
In this review we focus on macro-autophagy (in the following referred to as “autophagy”) whereby the formation of a double membrane vesicle, the autophagosome, leads to the engulfment of intracellular components such as proteins and organelles. Subsequently, the autophagosome cargo is degraded and recycled after fusion of the autophagosome with the lysosome (28).
The autophagy process is divided into the several steps: Induction of autophagy, initiation of isolation membrane, vesicle nucleation, elongation and finally fusion.
Regulation of autophagy/induction of autophagy
Autophagy is a cellular response to various stresses, especially starvation. The serine/threonine protein kinase mammalian target of rapamycin (mTOR) is the sensor of nutrition status and a key regulator of cellular metabolism, its inhibition in absence of nutrition, especially lack of amino acids, leads to inhibition of cell growth and induction of autophagy (30,31).
Under nutrient-rich conditions and growth factor signaling, mTOR is activated leading to autophagy inhibition via PI3K-/AKT-mTOR pathway (30,32). mTOR is also part of mTORC1 and mTORC2 complexes (33).
mTORC1 serves as one of the main amino-acid sensors in cell metabolism (30) and is indirectly activated by AKT-mediated inhibition of the mTOR negative regulator tuberous sclerosis complex (TSC) (32). The inhibition of TSC complex enables the small GTPase Rheb (Ras homolog enriched in brain) to activate mTORC1, after the small Rag GTPase recruited mTORC1 to the surface of the lysosome (34).
mTORC1 is thereby able to promote cell growth and inhibit the catabolic activity of autophagy by inhibition of Unc-51 like autophagy activating kinase (ULK1), a serine/threonine protein kinase responsible for autophagy initiation, and by inhibition of the transcription factor transcription factor EB (TFEB) (31,35,36). TFEB transcribes for lysosomal genes and autophagy genes and is involved in induction of autophagosome formation and autophagosome-lysosome fusion (37,38).
Under nutrient-rich conditions Rag recruits, besides mTORC1, cytosolic TFEB to the lysosomal membrane, where mTORC1 can then phosphorylate TFEB and prevent its nuclear translocation (32,34).
mTORC2 is not directly involved in autophagy regulation, but can influence autophagy inhibition through phosphorylation and activation of AKT (39). Therefore, mTORC2 enables the activation of mTORC1 by inhibition of TSC complex through AKT activation (32).
AMP-activated protein kinase (AMPK), a kinase responsible for the activation of autophagy, is the counterpart of mTOR in autophagy regulation. AMPK functions as an energy sensor and is stimulated by elevated AMP level to promote catabolic pathways for generation of energy (40).
AMPK and AKT interact with each other through mutual phosphorylation (41). AKT reduces AMPK phosphorylation at Thr-172, which is required for AMPK activity, by phosphorylation of AMPK at Ser-487/491 or by direct blocking of AMPK phosphorylation (41). Also AMPK is able to indirectly inhibit AKT activity (41).
Prolonged starvation induces AMPK, which then activates TSC2 by phosphorylation, leading to the inhibition of mTORC1 (42) and the activation of ULK1 (43,44).
Another important regulator of autophagic activity is the calcium-mediated activation of Ca2+/Calmodulin-dependent kinase kinase-β (CaMKK-β) (45). Reduction of amino acids content increases free cytosolic Ca2+ and promotes formation of Ca2+/Calmodulin complexes by inducing calcium efflux from intracellular stores such as the ER and the lysosomes into the cytosol (46). Ca2+/calmodulin activated CaMKK-β stimulates the autophagy inducer AMPK, leading to mTORC1 inhibition and ULK1 stimulation (45). In Addition, CaMKK-β can further induce the phosphatase calcineurin, which dephosphorylates TFEB and favors its nuclear translocation (47).
The ULK1 will complex with the autophagy genes transcribed from TFEB and then start the autophagy initiation (38,48).
Autophagosome formation
The ULK1 complex, the VPS34 complex and the ubiquitin-like conjugation systems ATG5-ATG12 and MAP1LC3-PE (microtubule associated protein light chain 3-phosphatydilethanolammine) will be the functional units of autophagosome formation (49).
Initiation and nucleation of the isolation membrane
The ULK1 complex and VPS34 (vacuolar protein sorting 34) complex, an autophagy specific class III PI3K complex, form a membranous cistern called isolation membrane (initially called “phagophore”) (50,51).
Under starvation, ULK1 is localized on the isolation membrane and forms, additionally, a complex with ATG13, FIP200 (focal adhesion kinase family interacting protein of 200 kDa) and ATG101 (50,52,53). The ULK1 complex recruits the VPS34 complex to the autophagy machinery and increases VPS34 activity resulting in enhanced phosphatidylinositol-3-phosphate (PI3P) production (48). PI3P accumulates at the ER membrane and promotes nucleation and growth of the autophagosome isolation membrane (54).
The VPS34 complex is formed by the autophagy-specific class III PI3K (VPS34), Beclin-1, ATG14L and VPS15 (51); its formation is disrupted by the inhibitory effect of the anti-apoptotic protein B-cell lymphoma-2 (Bcl-2), which blocks calcium channels efflux out of the ER, and inhibits the ULK1-dependent phosphorylation of Beclin-1 (55-57).
Elongation of the isolation membrane
The process of elongation and closure of the autophagosome isolation membrane depend on the two ubiquitin-like conjugation systems MAP1LC3-PE (LC3-II) and ATG5-ATG12 (58).
The cytosolic form of the ubiquitin like protein LC3 is cleaved at its carboxyl-terminus by the proteases ATG4 to form cytosolic LC3-I with an exposed c-terminal glycine (59,60). Thereby LC3-I can be conjugated at its c-terminal glycine with phosphatidylethanolamine, which is located on the isolation membrane, to form membrane-bound LC3-II (LC3-phosphatidylethanolamine/LC3-PE) (60). This process is mediated by the conjugation system ATG5-ATG12 (61).
The E1 enzyme ATG7 and the E2 enzyme ATG10 enable the conjugation system ATG5-ATG12, which is located in the isolation membrane and facilitates LC3-I to be bound on the membrane of the developing autophagosome (49,58).
The engulfment of cytosolic components principally acts not only as unspecific degradation mechanism but also as a selective up-take of intracellular cargo such as protein aggregates through a link between LC3-II and p62/SQSTM1 (62-64).
The ubiquitinated protein aggregates, and/or parts of the cytoplasm containing damaged organelles are surrounded by the isolation membrane that will form a double membrane-bound autophagosome by fusion of its terminal membrane parts.
Autophagosome-lysosome fusion
The autophagosome moves along microtubules to reach the lysosome (65) and delivers its cargo into the lysosomal lumen by fusion of the outer autophagosome membrane with lysosomal membrane.
The ability of the autophagosome to fuse with the lysosome depends on the small GTPases Rab7 (66) and is mediated by tethering factors and SNAREs (67,68).
At this point, the ATG conjugation systems are not necessary for autophagosome-lysosome fusion, but they are required for the degradation of the inner autophagosome membrane by lysosomal enzymes in the new formed autolysosome (69). The newly formed autolysosome can finally degrade the inner membrane and the cargo of the autophagosome. Thereby autophagy fulfills its physiological function of supplying energy and substrates to sustain cell homeostasis under starving conditions.
Autophagy in pathogenesis
As shown here before, several factors are enrolled in the regulation of autophagy process. The several steps of autophagosome formation and fusion with lysosome could be target of various impairments that could alter autophagy activity determining adverse effects in the intracellular environment. Additionally, impairment of autophagy could not rescue the cells during starvation.
Thus, it has been shown that dysregulations of autophagy are implicated in the development of many different diseases affecting the central nervous system and causing neurodegenerative disorders like Alzheimer and Parkinson, lysosomal storage diseases and tumorigenesis (3,4,26). Deletion of the essential ATG Beclin-1 promotes tumorigenesis in various cancer like breast cancer (5,6). In contrast to this, Beclin-1 overexpression and the consequently activation of autophagy inhibits tumorigenesis in breast carcinoma cells (26). Also the impairment of other genes involved in the positive regulation of autophagy such as PTEN (phosphatase and tensin homolog), which is an inhibitor of PI3K/Akt/mTOR pathway, can be crucial for cancer development (70).
Some cancer cells are able to build resistance against anti-tumor drugs by activating autophagy, resulting in cancer cell survival (71,72).
Autophagy in liver pathogenesis
Autophagy can act as a tumor suppressor by compensation of cellular distress or it can promote tumor growth by acting as a mechanism of cell survival (73).
Here, we report the involvement of autophagy in the development of pathologies correlated with liver.
It has been evidenced that autophagy is an important regulator of liver homeostasis. In particular, reticulophagy and mitophagy, even being autophagy processes, are acting independently and are highly increased in patients affected by severe steatosis (74). Furthermore, Fukushima et al. have shown that accumulation of p62 inclusion bodies in patients affected by non-alcoholic fatty liver disease (NAFLD) is correlated with macrophage infiltration, thus correlating the inflammatory response with ongoing autophagy process (75). Interestingly, Wang et al. discovered that the dietary intake of medium chain fatty acids in mice lower the lipotoxicity caused by high fat diet and mitigated type 2 diabetes and non-alcoholic steatohepatitis (NASH) via Rubicon-mediated autophagy (76). Our unpublished data showed an involvement of autophagy during NASH progression in mice and the correlation between Leptin and AMPK-mediated autophagy.
The autophagy process plays also a key role in alcohol-related liver disease. Injury of the hepatic tissue caused by alcohol in SNX10 knockout mice was determined by activation of autophagy. In particular, it was observed that the loss of SNX10 gene in mice determined the over-expression of autophagy markers LAMP-2A, Nrf2 and AMPK in alcohol-mediated liver steatosis (77). Furthermore, several studies noticed that alcohol consumption induce adipose tissue atrophy leading to autophagy impairment and block of tissue homeostasis, which lead to development of diseases correlated with development of pathologies affecting several tissue and organs, including liver (78).
HBV and HCV are responsible for chronic infection of hepatocytes. Tissue microarray analysis of human liver biopsy of patients infected by HBV and HCV revealed an up-regulation of MAP1LC3B expression in infected hepatocytes compared with non-infected liver cells (79). Interestingly, the up-regulation of autophagy process can be mediated by the expression of the HBV X protein, which binds to and activates phosphatidylinositol-3-kinase class 3 (PI3KC3). Thus, the viruses can use this catabolic process to enhance the duplication of viral DNA (80). Furthermore, this mechanism could be clarified by the ability of the HBV X protein to promote the nuclear translocation of high mobility group B1 (HMGB1) that could trigger the transcription of autophagy genes (81,82). Interestingly, an accumulation of p62 has been observed in patients with HCC involving chronic HBV infection and aflatoxin B1 (AFB1) exposure. These patients have shown a poor overall survival that could be correlated with the high expression of p62 (83). Once more, the HBV could use autophagy process to promote the degradation of tumor necrosis factor superfamily member 10 receptor (TNFSR10B/TRAILR1/DR5) in order to suppress the immune surveillance against virus-infected or transformed cells, thus inhibiting immune-mediated apoptosis (84). Interestingly, autophagy could still play a protective role against viral replication. The oxidative cellular stress caused by the HBV infection can induce the subunit of AMPK, PRKAA. Activation of AMPK promotes autolysosome-dependent degradation of HBV viral particles through stimulation of cellular ATP levels, which then leads to the depletion of autophagic vacuoles (85). Furthermore, our study has shown that HBV envelope proteins are responsible of ER stress induction in liver cancer cells in CB1-dependent manner (86). This mechanism could be responsible of promotion of autophagy as shown by Döring et al. and Lazar et al. (87,88), by the induction of the ER-associated degradation (ERAD) and the over-expression of Atg5-12/16L1 and Atg10/Atg3 complexes. Nonetheless, miRNAs could exert a key role by regulating autophagy in NASH. Several miRNAs of the maternally imprinted region at the chromosome 14q32.2 have been shown of being modulated in a NAFLD mouse model and their expression could exert an inhibitory effect on the expression of several targets including autophagic markers (89,90).
Autophagy and liver cancer
The development of liver diseases caused by high fat diet together with insulin-related pathologies, alcohol abuse and viral infection are responsible for activating tumorigenesis process in the liver, thus involving autophagy process. The first evidence of autophagy involvement in tumorigenesis and in hepatocarcinogenesis also, was shown by the development of mice expressing a heterozygous mutant form of Beclin-1. Those mice showed an impaired autophagy process and a high incidence of spontaneous tumors including liver cancer (91). The mutant heterozygous Beclin-1 was also responsible for increasing the frequency of spontaneous malignancies and accelerates the development of hepatitis B virus-induced premalignant lesions. Thus, showing that Beclin-1 is a haploinsufficient tumor suppressor (6).
Furthermore, Takamura et al. (7) showed that the deletion of ATG5 and ATG7 is responsible for the development of benign liver adenomas in mice characterized by autophagy impairment and accumulation of p62. Simultaneous deletion of p62 reduced the size of the Atg7−/− liver tumors. Other studies evidenced the involvement of p62 during liver tumorigenesis. In particular, oxidative stress determined by defective autophagy in tumor cells is characterized by accumulation of p62, reactive oxygen species, damaged mitochondria and ER chaperones. Sustained p62 expression resulting from autophagy defects was sufficient to alter NF-κB regulation and gene expression and to promote tumorigenesis (8). Wu et al. (92) found that increased autophagic activity promotes the ubiquitination and p62-mediated degradation of the oncogenic cyclin D1, which is high expressed in patients affected by HCC.
Aberrant autophagy is responsible for the over-expression of HGF in cirrhotic liver of rats. Thus, sustaining hepatocarcinogenesis via Met/JNK and Met/STAT3 signaling (93).
Liver non-parenchymal cells, including hepatic resident macrophage Kupffer cells, are also influenced by defects in the autophagy process. In autophagy-deficient macrophages, mitochondrial ROS mediated inflammation- and fibrosis-promoting effects by increasing IL1α/β production via enhancing NF-κB-associated pathways (94).
Recently, a protective role of thyroid hormone (TH) has been discovered during liver tumorigenesis. In detail, this study has shown that administration of TH is responsible for autophagy activation by promoting DAPK2 expression that mediates the phosphorylation of p62. Autophagy-mediated clearance of protein aggregates, in diethylnitrosamine-induced HCC mouse model, attenuates the hepatotoxicity and liver tumorigenesis (95).
In contrast to the protective role of autophagy against hepatocarcinogenesis, other studies have shown the implication of this catabolic process as tumor promoting mechanism. The long non-coding RNAs (lncRNA) have been implicated for autophagy-mediated liver carcinogenesis. In particular, the lncRNA HOTAIR resulted over-expressed in HCC. Its over-expression was responsible for the up-regulation of ATG5 and ATG7 genes, thus promoting tumor cell proliferation (96). Once more, the study of Umemura et al. (97) showed that p62 activity is required for the activation of NRF2 and mTORC1, the induction of c-Myc and to protect the HCC cells from the oxidative stress.
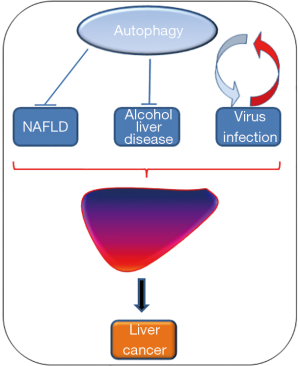
To summarize, the role exerted by autophagy to protect from oxidative stress, starvation and insult coming from viral infection, high fat diet and alcohol represent an important mechanism to protect the cells from stress and injury but tumor cells are also able to use this catabolic process to protect from stress and further proliferate (Figure 1).
Targeting autophagy in liver cancer
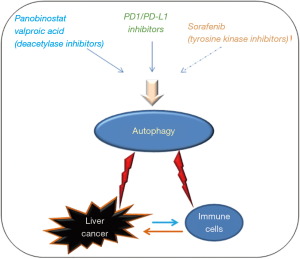
As described here above, the autophagy process requires the expression of several ATG genes and lysosomal genes. The autophagosome vesicle formation needs the interaction with ER and the involvement of several kinases that inhibit the mTOR/AKT pathway and activate the pro-autophagy factors. The final fusion with the lysosome is a complex mechanism that requires the action of lysosomal proteins. Based on this concern, autophagy is characterized by several druggable targets that could modulate autophagy status and promote anti-tumorigenic effects.
Our previous studies have shown that up-regulation of autophagy leads to cell death of HCC cells. Specifically, the treatment with the pan-deacetylase inhibitor panobinostat promoted an increase of the number of autophagy vesicles in HCC cells, the formation of the Beclin-1/MAP1LC3B/Atg12/UVRAG complex, the involvement of p53 and DRAM1 (DNA damage regulated autophagy modulator 1) and the final maturation of those, culminating into cell demise (10). This process could also be supported by the previous studies showing that panobinostat is able to trigger ER stress in HCC cells, which is also responsible for sustaining the autophagy process (98). Nonetheless, we have also shown that panobinostat caused the down-regulation of oncogenic miRNAs leading to the re-expression of several markers including autophagy proteins, e.g., Beclin-1 (99).
Further unpublished data have shown that autophagy process, specifically the expression of autophagy and ER stress related genes, could be regulated by the expression of CUX1 after treatment with deacetylase inhibitors. These studies evidenced that histone deacetylase inhibitors are capable to induce autophagy in HCC cells, which terminally causes cell demise.
The study of Zhang et al. (100) showed that transfection with a novel oncolytic adenovirus targeting Wnt signaling effectively inhibits cancer-stem like cell growth via metastasis, in HCC models. The transfection with the adenovirus sensitized the liver CSC-like to the treatment with classic substances like doxorubicin, which lead to autophagy related cell death. Valproic acid has shown to mediate the cellular internalization of doxorubicin in liver cancer HepG2 cells and to promote autophagy, reactive oxygen species and finally cell death. The activity was blocked by autophagy inhibitors (9).
The potent and selective Inhibitors of CLK1 have shown the peculiar ability to induce autophagy in mice. The up-regulation of autophagy process could protect the murine hepatocytes from the cytotoxic effect of acetaminophen (11).
Recent studies reported that the down-regulation of SIRT1 (NAD-dependent deacetylase sirtuin-1) signaling, a deacetylase responsible for cellular epigenetic reprogramming, underlies hepatic autophagy impairment in glycogen storage disease type Ia (101). The impairment of glucose-6-phosphatase-α (G6PC) can alter the metabolic programming of liver cells leading to inhibition of autophagy process and liver tumorigenesis (102).
The modulation of autophagy by treatment of HCC with sorafenib is still controversial. Some studies showed that sorafenib is able to promote autophagy-mediated cell death by up-regulation of autophagy markers, e.g., Beclin-1, by induction of ER stress and by suppression of the autophagy inhibitor mTORC1 and Akt (12,103-105). In contrast, it has been shown that PSMD10, together with Atg7, are markers of poor prognosis for patients affected by HCC. Treatment with sorafenib promotes the nuclear translocation of PSMD10 that promotes the transcription of Atg7 and induces autophagy resistance to sorafenib (106). Additionally, the inhibition of autophagy mediated by ADRB2 favors the stabilization of HIF-1a thus promoting sorafenib resistance in HCC cells (13).
Another recent study showed that the analysis of combined autophagic biomarkers like ULK1 and MAP1LC3B and their correlation with patient prognosis would better represent the dynamic stage of autophagy and it might provide a potential therapeutic way to target autophagy in HCC (107).
Recently, the discovery of the cell death signaling PD1 as a potential druggable target in several solid and blood malignancies (108-110) has given new possibilities for the treatment of cancer via the interference with the immune system. The potential of driving the immune response to mediate cell death of malignant cells could offer new perspectives for those kinds of tumors showing an involvement of the immune cells during their development stage. Liver cancer is a solid malignancy with high aggressiveness and metastatic potential which results in a poor prognosis for the patients. Triggering autophagy not only in the liver cancer cells but also in the immune cells involved in the tumor environment, e.g., T cells and macrophages, could improve the treatment of this fatal solid malignancy (Figure 2).
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Duve C, Pressman BC, Gianetto R, et al. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J 1955;60:604-17. [Crossref] [PubMed]
- Arstila AU, Trump BF. Studies on cellular autophagocytosis. The formation of autophagic vacuoles in the liver after glucagon administration. Am J Pathol 1968;53:687-733. [PubMed]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013;19:983-97. [Crossref] [PubMed]
- Seranova E, Connolly KJ, Zatyka M, et al. Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem 2017;61:733-49. [Crossref] [PubMed]
- Cicchini M, Chakrabarti R, Kongara S, et al. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 2014;10:2036-52. [Crossref] [PubMed]
- Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003;112:1809-20. [Crossref] [PubMed]
- Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011;25:795-800. [Crossref] [PubMed]
- Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009;137:1062-75. [Crossref] [PubMed]
- Saha SK, Yin Y, Kim K, et al. Valproic Acid Induces Endocytosis-Mediated Doxorubicin Internalization and Shows Synergistic Cytotoxic Effects in Hepatocellular Carcinoma Cells. Int J Mol Sci 2017.18. [PubMed]
- Di Fazio P, Waldegger P, Jabari S, et al. Autophagy-related cell death by pan-histone deacetylase inhibition in liver cancer. Oncotarget 2016;7:28998-9010. [Crossref] [PubMed]
- Sun QZ, Lin GF, Li LL, et al. Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers. J Med Chem 2017;60:6337-52. [Crossref] [PubMed]
- Tai WT, Shiau CW, Chen HL, et al. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis 2013;4. [Crossref] [PubMed]
- Wu FQ, Fang T, Yu LX, et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol 2016;65:314-24. [Crossref] [PubMed]
- Robainas M, Otano R, Bueno S, et al. Understanding the role of PD-L1/PD1 pathway blockade and autophagy in cancer therapy. Onco Targets Ther 2017;10:1803-7. [Crossref] [PubMed]
- Novikoff AB, Beaufay H, Duve C. Electron microscopy of lysosomerich fractions from rat liver. J Biophys Biochem Cytol 1956;2:179-84. [Crossref] [PubMed]
- Essner E, Novikoff AB. Human hepatocellular pigments and lysosomes. J Ultrastruct Res 1960;3:374-91. [Crossref] [PubMed]
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol 1962;12:198-202. [Crossref] [PubMed]
- Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol 1966;28:435-92. [Crossref] [PubMed]
- Klionsky DJ. Autophagy revisited: A conversation with Christian de Duve. Autophagy 2008;4:740-3. [Crossref] [PubMed]
- Deter RL, Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol 1967;33:437-49. [Crossref] [PubMed]
- Mitchener JS, Shelburne JD, Bradford WD, et al. Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am J Pathol 1976;83:485-92. [PubMed]
- Pfeifer U, Strauss P. Autophagic vacuoles in heart muscle and liver. A comparative morphometric study including circadian variations in meal-fed rats. J Mol Cell Cardiol 1981;13:37-49. [Crossref] [PubMed]
- Takeshige K, Baba M, Tsuboi S, et al. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992;119:301-11. [Crossref] [PubMed]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993;333:169-74. [Crossref] [PubMed]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011;27:107-32. [Crossref] [PubMed]
- Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999;402:672-6. [Crossref] [PubMed]
- Besterman JM, Low RB. Endocytosis: A review of mechanisms and plasma membrane dynamics. Biochem J 1983;210:1-13. [Crossref] [PubMed]
- Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell 2004;6:463-77. [Crossref] [PubMed]
- Tekirdag K, Cuervo AM. Chaperone-mediated autophagy and endosomal microautophagy: Joint by a chaperone. J Biol Chem 2018;293:5414-24. [Crossref] [PubMed]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014;24:400-6. [Crossref] [PubMed]
- Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009;20:1981-91. [Crossref] [PubMed]
- Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol 2015;25:545-55. [Crossref] [PubMed]
- Loewith R, Jacinto E, Wullschleger S, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 2002;10:457-68. [Crossref] [PubMed]
- Napolitano G, Ballabio A. TFEB at a glance. J Cell Sci 2016;129:2475-81. [Crossref] [PubMed]
- Vega-Rubin-de-Celis S, Peña-Llopis S, Konda M, et al. Multistep regulation of TFEB by MTORC1. Autophagy 2017;13:464-72. [Crossref] [PubMed]
- Wong PM, Puente C, Ganley IG, et al. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy 2013;9:124-37. [Crossref] [PubMed]
- Sardiello M, Palmieri M, Di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473-7. [Crossref] [PubMed]
- Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429-33. [Crossref] [PubMed]
- Sarbassov DD, Guertin DA, Ali SM, et al. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005;307:1098-101. [Crossref] [PubMed]
- Villanueva-Paz M, Cotán D, Garrido-Maraver J, et al. AMPK Regulation of Cell Growth, Apoptosis, Autophagy, and Bioenergetics. EXS 2016;107:45-71. [Crossref] [PubMed]
- Zhao Y, Hu X, Liu Y, et al. ROS signaling under metabolic stress: cross-talk between AMPK and AKT pathway. Mol Cancer 2017;16:79. [Crossref] [PubMed]
- Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003;115:577-90. [Crossref] [PubMed]
- Lee JW, Park S, Takahashi Y, et al. The association of AMPK with ULK1 regulates autophagy. PLoS One 2010;5. [Crossref] [PubMed]
- Yao Y, Jones E, Inoki K. Lysosomal Regulation of mTORC1 by Amino Acids in Mammalian Cells. Biomolecules 2017.7. [PubMed]
- Ghislat G, Patron M, Rizzuto R, et al. Withdrawal of essential amino acids increases autophagy by a pathway involving Ca2+/calmodulin-dependent kinase kinase-β (CaMKK-β). J Biol Chem 2012;287:38625-36. [Crossref] [PubMed]
- Kondratskyi A, Yassine M, Kondratska K, et al. Calcium-permeable ion channels in control of autophagy and cancer. Front Physiol 2013;4:272. [Crossref] [PubMed]
- Medina DL, Ballabio A. Lysosomal calcium regulates autophagy. Autophagy 2015;11:970-1. [Crossref] [PubMed]
- Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013;15:741-50. [Crossref] [PubMed]
- Mizushima N, Yoshimori T, Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2003;35:553-61. [Crossref] [PubMed]
- Ganley IG, Du Lam H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009;284:12297-305. [Crossref] [PubMed]
- Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 2013;9:1983-95. [Crossref] [PubMed]
- Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008;181:497-510. [Crossref] [PubMed]
- Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009;5:649-62. [Crossref] [PubMed]
- Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett 2010;584:1302-12. [Crossref] [PubMed]
- Høyer-Hansen M, Bastholm L, Szyniarowski P, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 2007;25:193-205. [Crossref] [PubMed]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005;122:927-39. [Crossref] [PubMed]
- Park JM, Seo M, Jung CH, et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 2018;14:584-597. [Crossref] [PubMed]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep 2008;9:859-64. [Crossref] [PubMed]
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000;19:5720-8. [Crossref] [PubMed]
- Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J Biol Chem 2004;279:47704-10. [Crossref] [PubMed]
- Otomo C, Metlagel Z, Takaesu G, et al. Structure of the human ATG12~ATG5 conjugate required for LC3 lipidation in autophagy. Nature structural & molecular biology 2013;20:59-66. [Crossref] [PubMed]
- Noda NN, Kumeta H, Nakatogawa H, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes to Cells 2008;13:1211-8. [Crossref] [PubMed]
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007;282:24131-45. [Crossref] [PubMed]
- Ichimura Y, Kumanomidou T, Sou Y-s, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 2008;283:22847-57. [Crossref] [PubMed]
- Fass E, Shvets E, Degani I, et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem 2006;281:36303-16. [Crossref] [PubMed]
- Gutierrez MG, Munafó DB, Berón W, et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004;117:2687-97. [Crossref] [PubMed]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012;151:1256-69. [Crossref] [PubMed]
- Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell 2014;25:1327-37. [Crossref] [PubMed]
- Tsuboyama K, Koyama-Honda I, Sakamaki Y, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016;354:1036-41. [Crossref] [PubMed]
- Rosenfeldt MT, O'Prey J, Flossbach L, et al. PTEN deficiency permits the formation of pancreatic cancer in the absence of autophagy. Cell Death Differ 2017;24:1303-4. [Crossref] [PubMed]
- Buchser WJ, Laskow TC, Pavlik PJ, et al. Cell-mediated autophagy promotes cancer cell survival. Cancer Res 2012;72:2970-9. [Crossref] [PubMed]
- Shuhua W, Chenbo S, Yangyang L, et al. Autophagy-related genes Raptor, Rictor, and Beclin-1 expression and relationship with multidrug resistance in colorectal carcinoma. Hum Pathol 2015;46:1752-9. [Crossref] [PubMed]
- Singh SS, Vats S, Chia AY-Q, et al. Dual role of autophagy in hallmarks of cancer. Oncogene 2018;37:1142-58. [Crossref] [PubMed]
- Pang L, Liu K, Liu D, et al. Differential effects of reticulophagy and mitophagy on nonalcoholic fatty liver disease. Cell Death Dis 2018;9:90. [Crossref] [PubMed]
- Fukushima H, Yamashina S, Arakawa A, et al. The formation of p62-positive inclusion body is associated with macrophage polarization in non-alcoholic fatty liver disease. Hepatol Res 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Wang ME, Singh BK, Hsu MC, et al. Increasing Dietary Medium-Chain Fatty Acid Ratio Mitigates High-fat Diet-Induced Non-Alcoholic Steatohepatitis by Regulating Autophagy. Sci Rep 2017;7:13999. [Crossref] [PubMed]
- You Y, Li WZ, Zhang S, et al. SNX10 mediates alcohol-induced liver injury and steatosis by regulating the activation of chaperone-mediated autophagy. J Hepatol 2018;69:129-41. [Crossref] [PubMed]
- Li Y, Ding WX. Adipose tissue autophagy and homeostasis in alcohol-induced liver injury. Liver Res 2017;1:54-62. [Crossref] [PubMed]
- Yeganeh B, Rezaei Moghadam A, Alizadeh J, et al. Hepatitis B and C virus-induced hepatitis: Apoptosis, autophagy, and unfolded protein response. World J Gastroenterol 2015;21:13225-39. [Crossref] [PubMed]
- Sir D, Ann DK, Ou JH. Autophagy by hepatitis B virus and for hepatitis B virus. Autophagy 2010;6:548-9. [Crossref] [PubMed]
- Fu S, Wang J, Hu X, et al. Crosstalk between hepatitis B virus X and high-mobility group box 1 facilitates autophagy in hepatocytes. Mol Oncol 2018;12:322-38. [Crossref] [PubMed]
- Cheng LS, Li J, Liu Y, et al. HMGB1-induced autophagy: a new pathway to maintain Treg function during chronic hepatitis B virus infection. Clin Sci (Lond) 2017;131:381-94. [Crossref] [PubMed]
- Xiang X, Qin HG, You XM, et al. Expression of P62 in hepatocellular carcinoma involving hepatitis B virus infection and aflatoxin B1 exposure. Cancer Med 2017;6:2357-69. [Crossref] [PubMed]
- Shin GC, Kang HS, Lee AR, et al. Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy 2016;12:2451-66. [Crossref] [PubMed]
- Xie N, Yuan K, Zhou L, et al. PRKAA/AMPK restricts HBV replication through promotion of autophagic degradation. Autophagy 2016;12:1507-20. [Crossref] [PubMed]
- Montalbano R, Honrath B, Wissniowski TT, et al. Exogenous hepatitis B virus envelope proteins induce endoplasmic reticulum stress: involvement of cannabinoid axis in liver cancer cells. Oncotarget 2016;7:20312-23. [Crossref] [PubMed]
- Döring T, Zeyen L, Bartusch C, et al. Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does not Require Atg8/LC3 Lipidation for Viral Maturation. J Virol 2018.92. [PubMed]
- Lazar C, Macovei A, Petrescu S, et al. Activation of ERAD pathway by human hepatitis B virus modulates viral and subviral particle production. PLoS One 2012;7. [Crossref] [PubMed]
- Okamoto K, Koda M, Okamoto T, et al. A Series of microRNA in the Chromosome 14q32.2 Maternally Imprinted Region Related to Progression of Non-Alcoholic Fatty Liver Disease in a Mouse Model. PLoS One 2016;11. [Crossref] [PubMed]
- Di Fazio P, Wissniowski TT. Comment on “A series of microRNA in the chromosome 14q32.2 maternally imprinted region related to progression of non-alcoholic fatty liver disease in a mouse model”. Hepatoma Res 2016;2:205-6. [Crossref]
- Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003;100:15077-82. [Crossref] [PubMed]
- Wu SY, Lan SH, Wu SR, et al. Hepatocellular carcinoma-related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Li J, Hu SB, Wang LY, et al. Autophagy-dependent generation of Axin2+ cancer stem-like cells promotes hepatocarcinogenesis in liver cirrhosis. Oncogene 2017;36:6725-37. [Crossref] [PubMed]
- Sun K, Xu L, Jing Y, et al. Autophagy-deficient Kupffer cells promote tumorigenesis by enhancing mtROS-NF-kappaB-IL1alpha/beta-dependent inflammation and fibrosis during the preneoplastic stage of hepatocarcinogenesis. Cancer Lett 2017;388:198-207. [Crossref] [PubMed]
- Chi HC, Chen SL, Tsai CY, et al. Thyroid hormone suppresses hepatocarcinogenesis via DAPK2 and SQSTM1-dependent selective autophagy. Autophagy 2016;12:2271-85. [Crossref] [PubMed]
- Yang L, Zhang X, Li H, et al. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol Biosyst 2016;12:2605-12. [Crossref] [PubMed]
- Umemura A, He F, Taniguchi K, et al. p62, Upregulated during Preneoplasia, Induces Hepatocellular Carcinogenesis by Maintaining Survival of Stressed HCC-Initiating Cells. Cancer Cell 2016;29:935-48. [Crossref] [PubMed]
- Montalbano R, Waldegger P, Quint K, et al. Endoplasmic reticulum stress plays a pivotal role in cell death mediated by the pan-deacetylase inhibitor panobinostat in human hepatocellular cancer cells. Transl Oncol 2013;6:143-57. [Crossref] [PubMed]
- Henrici A, Montalbano R, Neureiter D, et al. The pan-deacetylase inhibitor panobinostat suppresses the expression of oncogenic miRNAs in hepatocellular carcinoma cell lines. Mol Carcinog 2015;54:585-97. [Crossref] [PubMed]
- Zhang J, Lai W, Li Q, et al. A novel oncolytic adenovirus targeting Wnt signaling effectively inhibits cancer-stem like cell growth via metastasis, apoptosis and autophagy in HCC models. Biochem Biophys Res Commun 2017;491:469-77. [Crossref] [PubMed]
- Cho JH, Kim GY, Pan CJ, et al. Downregulation of SIRT1 signaling underlies hepatic autophagy impairment in glycogen storage disease type Ia. PLoS Genet 2017;13. [Crossref] [PubMed]
- Cho JH, Kim GY, Mansfield BC, et al. Hepatic glucose-6-phosphatase-alpha deficiency leads to metabolic reprogramming in glycogen storage disease type Ia. Biochem Biophys Res Commun 2018;498:925-31. [Crossref] [PubMed]
- Shi YH, Ding ZB, Zhou J, et al. Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy 2011;7:1159-72. [Crossref] [PubMed]
- Zhai B, Hu F, Jiang X, et al. Inhibition of Akt reverses the acquired resistance to sorafenib by switching protective autophagy to autophagic cell death in hepatocellular carcinoma. Mol Cancer Ther 2014;13:1589-98. [Crossref] [PubMed]
- Ling S, Song L, Fan N, et al. Combination of metformin and sorafenib suppresses proliferation and induces autophagy of hepatocellular carcinoma via targeting the mTOR pathway. Int J Oncol 2017;50:297-309. [Crossref] [PubMed]
- Luo T, Fu J, Xu A, et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy 2016;12:1355-71. [Crossref] [PubMed]
- Wu DH, Wang TT, Ruan DY, et al. Combination of ULK1 and LC3B improve prognosis assessment of hepatocellular carcinoma. Biomed Pharmacother 2018;97:195-202. [Crossref] [PubMed]
- Annibali O, Crescenzi A, Tomarchio V, et al. PD-1 /PD-L1 checkpoint in hematological malignancies. Leuk Res 2018;67:45-55. [Crossref] [PubMed]
- Bertucci F, Finetti P, Mamessier E, et al. PDL1 expression is an independent prognostic factor in localized GIST. Oncoimmunology 2015;4. [Crossref] [PubMed]
- Bansal P, Osman D, Gan GN, et al. Recent Advances in Immunotherapy in Metastatic NSCLC. Front Oncol 2016;6:239. [PubMed]
Cite this article as: Di Fazio P, Matrood S. Targeting autophagy in liver cancer. Transl Gastroenterol Hepatol 2018;3:39.


