SUMOylation and phosphorylation cross-talk in hepatocellular carcinoma
Hepatocellular carcinoma (HCC) and post-translation modification
HCC is a primary malignancy of the liver and the third leading cause of cancer deaths worldwide. HCC occurs predominantly in patients with underlying chronic liver disease and cirrhosis progressing with local expansion, intrahepatic spread, and distant metastases (1). Asia and Africa are considered the countries with the highest peak of HCC incidence, and at the same time appeared to be afflicted by an impressive prevalence of hepatitis B and C, well-known to predispose to HCC development. Nonalcoholic fatty liver disease (NAFLD) or nonalcoholic steatohepatitis (NASH)-induced cirrhosis is considered a growing health problem. Several research groups reported that NASH is commonly developed under obesity, type 2 diabetes, dyslipidemia and hypertension conditions. The obesity epidemic is a global phenomenon, but it is worse in the United States that in other developed countries (2). These findings suggest that more patient-focused care should be provided during the end-stage of liver disease and HCC.
A broad number of protein post-translational modification (PTM) includes several critical signaling events that occur during neoplastic transformation in liver, colon, breast, gastric and pancreatic tissues (3). More than 200 different PTMs were identified in both prokaryotic and eukaryotic cells, that occur in almost all proteins and increase their functional diversity, including ubiquitination, phosphorylation, glycosylation, acetylation, SUMOylation and methylation (4). New tools development to identify PTMs have permitted tremendous advancement in understanding how these important processes are regulated and how they influence protein function in liver pathophysiology.
Role of SUMOylation in HCC
SUMOylation machinery
SUMOylation has recently emerged as a key PTM that regulates protein stability, protein-protein interactions, trafficking and transcriptional activity (5). Similar to ubiquitination, that uses an equivalent reaction scheme and enzyme classes, to conjugate ubiquitin, SUMOylation acts through a small ubiquitin-like modifiers (SUMOs). The SUMO acceptor site is often situated within a consensus motif whose core sequence is Ψ-K-x-D/E, where Ψ represents a large hydrophobic residue, K is the modified Lys residue, x is any residue, and D/E represents asparagin (Asp) or glutamine (Glu) (6). More than 1,000 proteins have been identified as potential SUMO-conjugation (SUMOylation) targets. SUMO polypeptide is first cleaved at the C-terminal domain, then activated by binding to a heterodimeric E1 enzyme (SUMO activating enzyme composed of subunits 1 and 2 or SAE1/SAE2), then transferred to the E2 conjugating enzyme (UBC9) and finally E3 enzymes (RanBP2, PIAS, and the polycomb group protein Pc2) enhance the transfer of SUMO from E2 to specific substrates (7) (Figure 1). SUMO-specific proteases (SENPs) process SUMO precursors to mature forms and also remove SUMO from its targets (8). In humans, three functional SUMO isoforms (SUMO-1, SUMO-2, and SUMO-3) are encoded in the genome, with SUMO-2/3 sharing a high degree of identity (9). They can be conjugated to distinct and overlapping sets of proteins and play essential roles in various cellular processes. SUMOylation changes substrate protein properties, in particular by favoring the recruitment of SUMO-binding partners (6,7). SUMOylation is sensitive to various stresses that regulate the activity of the SUMO pathway’s enzymes.
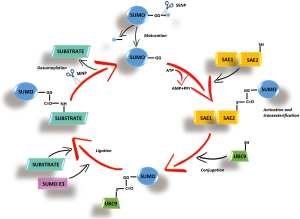
Recently, SUMO-1 was found to be highly expressed in HCC cell lines and histological HCC samples, while its expression level appeared drastically low in non-neoplastic liver tissues (10). Moreover, the expression of SAE1/2 has been found significantly upregulated in cancer tissues of HCC patients (11). Intriguingly, in liver cancer patients the survival rate is strongly correlated with SUMO-2 expression. In addition, UBC9, the sole E2 enzyme in SUMOylation machinery, has been described overexpressed in HCC during SUMO modification (12), while SENP2, that controls the deSUMOylation process, inhibits the proliferation of HCC cells (13,14). Moreover, SUMOylation appears to play a key role in HCC multidrug resistance (15).
SUMOylation and gene transcription
Recent human SUMO proteomics studies have shown that nearly 300 sequence-specific DNA-binding transcription factors (TFs), that act as activators or repressors of transcription by RNA polymerase II (RNAP II), are SUMO conjugates, which represents over 50% of all TFs in this category, indicating that they are particularly subject of regulation by SUMO (16,17). SUMO can impair transcription by promoting the recruitment of transcriptional corepressor complexes or by interfering with transcription-promoting PTMs, such as acetylation and phosphorylation (18). TF SUMOylation is most often associated with reduced target gene expression, which can be mediated by enhanced interactions with corepressors or by interference with protein modifications that promote transcription (19,20). However, it is becoming evident that SUMOylation also regulates gene expression by controlling the levels of TFs that are associated with chromatin. This can occur directly, by influencing DNA-binding ability or promoting clearance of TFs from chromatin, or indirectly, by regulating TFs abundance or localization (18).
MATα2
In mammals, two different genes, MAT1A (expressed mostly in liver) and MAT2A (expressed in all extra-hepatic tissues), encode for MAT isoenzymes. essential cellular enzymes responsible for S-adenosylmethionine biosynthesis (SAMe). SAMe is the link of three key metabolic pathways, polyamine synthesis, transmethylation and transsulfuration (21) and is the best known methyl donor. Although, hepatic SAMe depletion and exogenous SAMe administration affect PTMs, such as SUMOylation and phosphorylation (12,22). MAT1A is a marker of normal differentiated liver, whereas MAT2A is widely distributed, and it is down-regulated in chronic liver disease and often silenced in HCC. Several studies showed a switch from MAT1A to MAT2A expression in human HCC, which facilitates cancer cell growth (23). We previously demonstrated that the expression level of UBC9 is higher in HCC and when hepatic SAMe level falls. SAMe administration lowered SUMOylation reducing UBC9 protein level in Mat1a KO mice (12). Recently, UBC9 has been shown to be a positive regulator of B-cell lymphoma 2 (Bcl-2) expression promoting cell survival rather than by driving cell proliferation as critical step in tumor development by unknown mechanism (22). Intriguingly, Bcl-2 expression is regulated by MATα2 at multiple levels. MATα2 can directly bind Bcl-2 promoter to induce its transcription and also directly interact with Bcl-2 to enhance its protein stability. The effect of MATα2 on Bcl-2 requires UBC9 to control MATα2’s stability that is influenced by SUMOylation at K340, K372 and K394 (12). The cross-talk between SUMOylated MATα2 and Bcl-2 can confer survival benefit to the cancer cell.
Nrf2
The pathophysiological changes that control the liver cirrhosis progression to HCC are under oxidative stress influence. The liver is susceptible to organ toxicity by xenobiotic and other chemicals biotransformation and detoxification because high level of oxidative stress, by reactive intermediates production, is generated (24). Nuclear erythroid 2-related factor 2 (Nrf2) is an important TF that plays a key role in adaptive responses to oxidative stress. Nrf2 directly binds the antioxidant response element (ARE) sequences of numerous antioxidant and cytoprotective genes to drive the cellular response (25). Glutathione (GSH) is a tripeptide comprised of three amino acids (cysteine, glutamic acid, and glycine) present in most mammalian tissue that acts as an antioxidant, a free radical scavenger and a detoxifying agent. In addition, it is synthesized by glutamate-cysteine ligase (GCL) and GSH synthase enzymatic complex to defend the cell against oxidative stress (26). Bacterial lipopolysaccharide (LPS) is the major molecular component of the outer membrane of Gram-negative bacteria that provides a physical barrier between the bacteria its surroundings. LPS is well-known to cause liver failure lowering the hepatic GSH level through Nrf2 SUMOylation inhibition (27). In fact, reduced levels of Nrf2 and v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G (MafG) SUMOylation, which is required for their heterodimerization and subsequent binding and trans-activation of the ARE element present in the promoter region of Gclc and Gclm genes, are important for their expression (26,27). Moreover, liver fibrosis study elucidated the mechanism by which Nrf2 and MafG SUMOylation enhances their heterodimerization and increases Gclc expression that is essential to maintain the hepatic stellate cells under quiescent state preventing liver failure (26).
p53
The tumor-suppressor protein p53, encoded by TP53 gene, inhibits tumorigenesis by inducing cell-cycle arrest, senescence, and apoptosis. In addition to apoptosis, cytoplasmic p53 is able to mediate other non-transcriptional functions, such as autophagy and inhibition of glucose metabolism (28). Most research relating to p53 SUMOylation focused on the effects of SUMO on p53 transcriptional activities, that can either enhances or reduces its activity (29). Similarly, MDM2, the main ubiquitin E3 ligase for p53, is SUMOylated at N-terminal domain between amino acids 40–59 sequence and required to induce MDM2 activity (29). In addition, MDM2 protein degradation is under its self-ubiquitination control. However, MDM2 SUMOylation prevents protein degradation competing with its self-ubiquitination, while de-SUMOylation induces MDM2 self-ubiquitination. Previously, SUMO-1 has been found to enhance apoptosis in HepG2 cells via p53 SUMOylation. In other hands, the apoptosis rate in the cells transfected with pwtp53 + pMDM2 + pSUMO-1, appeared significantly higher compared to cells transfected with pwtp53 + pMDM2. Also, SUMO-1/p53 complex has been found to prevent MDM2-dependent p53 protein degradation (30).
SKI is a protooncogene that has been described able to transform avian fibroblasts in vitro (31). The oncogenic activity of this intriguing player was explored and explained by numerous molecular mechanisms. For example, SKI appears to be a key protein on TGF-β signaling because it is able to physically interact with Smad proteins including Smad2, Smad3, and Smad4, and this prevents their transcriptional activation (32). It has also been described to form a complex with retinoblastoma tumor suppressor that inhibits the transcriptional repression (33). In addition, SKI protein induces the thioester bond formation between UBC9 and SUMO-1 and also promotes UBC9 SUMOylation itself (34). Knipscheer et al. reported that UBC9 SUMOylation influences the substrate SUMOylation (35), suggesting a plausible mechanism by which SKI may indirectly induce thioester bond formation providing more SUMO donor, where SKI appears as a SUMO E3 for UBC9. Moreover, SKI is capable to regulate MDM2 SUMOylation interacting with and regulating UBC9, that finally induces MDM2 activity and lowers p53 protein (34). Particularly, SKI promotes MDM2 SUMOylation via PIAS1 and PIAS3 that increases MDM2 level, therefore induces p53 protein degradation through ubiquitination and proteasome system.
HIF-1α and Cbx4
Hypoxia-inducible factor-1 (HIF-1) is an important TF involved on cellular adaptive response to hypoxia and is commonly described to be linked to the solid tumor microenvironment. One of the most important subunit oxygen-sensitive of HIF-1 is HIF-1α, well-known to be regulated by PTMs (36). More specifically, SUMOylation affects HIF-1α transcriptional activity altering its interaction with co-modulators that could be vary from cell type to cell type. More intriguingly, the complex formation between Cbx4, a polycomb group protein (SUMO E3 ligase), and PIASy (E3 SUMO-protein ligase) mediates HIF-1α SUMOylation that regulates its protein stability and transcriptional activity in HCC cell lines (37).
Analysis of a large cohort of HCC tissues reported that Cbx4 is highly expressed and positively correlated with poor overall survival of HCC patients (37,38). Interestingly, Cbx4 is the sole Cbx family members that induces angiogenesis promoting the expression of hypoxia-induced vascular endothelial growth factor (VEGF) in HCC cells via induced-HIF-1α SUMOylation. This occurs on its two SUMO-interacting motifs located at K391 and K477 residues, which finally increases the transcriptional activity of HIF-1α. In fact, mutating HIF-1α at K391 and K477 residues has no effect on PIASy regulatory function may be because Cbx4 and PIASy mediate HIF-1α SUMOylation at different lysine (37) or other specific SUMO E3 ligases generate additional SUMOylation patterns that influence the stability and transcriptional activity of HIF-1α. However, the potential HIF-1α SUMOylated binding sites by PIASy and other SUMO E3 ligases remain to be investigated. In addition, proteomic experiments revealed that non-SUMOylated HIF-1α and HIF-1α proteins interact with different partners when SUMOylation is induced by Cbx4 (37), then it is extremely important to explore their role on HIF-1α stability and function. In addition, to develop novel compounds targeting Cbx4 SUMO E3 ligase activity and its interaction with HIF-1α, could move forward in fight against tumor angiogenesis and its need for oxygen and nutrients.
SUMOylation and protein stability
Numerous research groups reported that cellular signaling is important to modulate stability and accessibility of proteins. The most important pathway is the ubiquitination that leads to proteasome degradation that could also be caused by losing the lysine acceptor for SUMOylation (39). This event supports the concept that these two protein modifications are closely interplaying. However, the biological consequences of SUMOylation and ubiquitination are quite different. Whereas ubiquitin primarily mediates protein degradation, SUMO serves several different purposes as well as protein stabilization by antagonizing ubiquitination (40). Intriguingly, in the same cases the SUMO-1 attachment to its target proteins protects against ubiquitination and lowers proteasomal degradation (30).
β-catenin
SUMO proteases (SENP) are known to be responsible of SUMO remotion from SUMOylated proteins, and their expression is described to be deregulated in many cancers (14,41). Specifically, SENP2 was found to be down-regulated in HCC tissues, while its overexpression is believed to prevent growth and colony formation of HCC cells. In contrast, SENP2 knockdown leads cancer cell growth (14).
WW domain-containing oxidoreductase (WWOX) is a novel oncosuppressor gene that appears to be downregulated in various cancers including HCC (42). WWOX seems to be involved on Wnt/β-catenin pathway inhibition by an associated mechanism with Disheveled protein (Dvl), an important molecule for β-catenin protein stabilization (43). WWOX has been described as an important downstream modulator of SENP2 tumor suppressor function, and a key player on SENP2-β-catenin asset that controls liver cancer growth. Basically, SENP2 negatively regulates Wnt/β-catenin pathway transcriptional activity by inducing WWOX transcription, thereby resulting in β-catenin protein destabilization (13,14). In contrast, WWOX silencing reverses the effect of SENP2 on β-catenin stability and cancer cells growth (13). In addition, β-catenin stability markedly dropped when SENP2 is overexpressed, while SENP2 catalytic mutant had no effects suggesting that this phenomena is SENP2 de-SUMOylation activity-dependent (14).
FOXA2
The forkhead box protein A2 (FOXA2) is a member of FoxA subfamily that plays a key role in glucose and lipid homeostasis controlling several genes expression in metabolically active tissues such as, liver, pancreatic α and β cells and adipocytes (44). Casein kinase I (CKI) and protein kinase B (AKT) control FOXA2 expression and activity phosphorylating its transactivation domain located at the N-terminus (45). However, these phosphorylation sites are not required for FOXA2 transcriptional activity. More recently, IKKα has been described to phosphorylate FOXA2 on serine 107 (S107) and 111 (S111) residues that inhibits its transcriptional activity, derepressed FOXA2 target genes and promoted liver cancer growth (46). Nevertheless, FOXA2 was found to be SUMOylated on lysine 6 (K6) residues, located within a consensus SUMOylation site, that is required for the stability of FOXA2 protein. Mutating SUMO acceptor lysine to arginine (R), completely abolished both in vivo and in vitro SUMOylation. Specifically, FOXA2K6R mutant protein stability markedly decreased without affecting its mRNA level, while SUMO-1 in-frame fusion construct promoted stability on unstable FOXA2K6R mutant protein (lack of SUMOylation conjugation). In addition, SUMO ligase PIAS1 increased FOXA2 level promoting FOXA2 SUMOylation, whereas the SUMO ligase dominant negative (PIAS1 deficient) lowered both SUMOylation and basal level of non-SUMOylated FOXA2 (47). Despite the importance of SUMOylation for FOXA2 protein stability, only a small fraction of FOXA2 is SUMOylated at basal level even though the non-SUMOylated FOXA2 appeared to be stable. Accordingly, non-SUMOylated FOXA2, derived from deSUMOylation of previously SUMOylated FOXA2 that remain stable whereas FOXA2K6R, is incapable to be SUMOylated and then it is destabilized (47).
p65
Abnormal activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway is closely related to hepatocarcinogenesis and HBV-related HCC (48,49). Concurrently, high level of NF-κB signaling and activity promote migration and invasive abilities of liver cancer cells (50). Moreover, NF-κB pathway is involved on reduced radiotherapeutics and chemotherapeutics sensitivity of HCC cells (51). In contrast, reducing NF-κB pathway prevents invasiveness and malignancy of human HCC cells and reduces carcinogenesis in vivo (52).
The human NF-κB family includes five different DNA-binding subunits: RelA/p65, c-Rel, RelB, p50 and p52. All NF-κB DNA-binding subunits have an N-terminal Rel homology domain (RHD), by which they set up dimerization, interaction with the inhibitor of κB (IκB) proteins, nuclear translocation and DNA binding (53). Only p65, RelB and cRel contain the C-terminal transactivation domains that are required for gene transcription ability. More specifically, p65 contains two potent transactivation domains within its C-terminus protein sequence that are responsible of its strongest capability to be activator of most genes containing κB sites in their sequences (53). p65 has been described as a critical key regulatory protein on NF-κB transcriptional activity and is associated with liver cancer development. When p65 is localized into the nucleus appears to be under its active form, that controls a large number of genes transcription in response to inflammatory stimuli (53,54). Accordingly, p65 protein stability regulation plays an important role in hepatitis-related HCC development. p65 has been previously found to be SUMOylated by exogenous SUMO-3 in HEK 293T cells and mouse 3T3 fibroblast cells (55). In addition, silencing of SUMO-2/3 promoted TNF-α-induced transcriptional activity of p65 in Hela cells (56). Recently, Liu et al. speculated that p65 and SUMO-2/3 interaction represents a novel mechanism by which p65 protein stability is regulated suppressing its degradation through ubiquitin-proteasome system in the cytoplasm (57). In details, p65 SUMOylation could be mediated by SUMO proteins that recruit an inhibitor of p65 in nucleus may be via SUMO-2/3 that has been described to inhibit nuclear import of p65 (58). Subsequentially, SUMO-2 or SUMO-3 induced the cytoplasmic level of p65 and may suppress its transcriptional activity (57,59).
Role of phosphorylation in HCC
Protein phosphorylation
Protein phosphorylation is a central signaling event in eukaryotes and is regulated by protein kinases that catalyze the transfer of the γ-phosphoryl group of a nucleoside triposphate [usually adenosine triphosphate (ATP)] to an amino acid hydroxyl group (commonly serine, threonine, or tyrosine) of a protein substrate. It is an important cellular regulatory mechanism as many enzymes and receptors are activated/deactivated by phosphorylation and dephosphorylation events (60). The most common sites of phosphorylation are serine, threonine and tyrosine in mammalian proteins (61). Protein phosphorylation has become a central focus of drug discovery as the result of the identification and validation of promising therapeutic targets such as protein kinases, protein phosphatases, and phosphoprotein binding domains.
Regulation of protein-protein interactions and protein stability by site-specific phosphorylation
Phosphorylation on serine or threonine residues can be crucial for regulating protein-protein interactions and protein stability. A well-known example is that of the famous tumor suppressor, p53 that plays an important role in the cellular response to stress such as DNA damage and hypoxia. Loss of p53 is associated with genomic instability and tumor development (62). Phosphorylation of p53 at serine 15, serine 20 and serine 33 are involved in preventing interaction of p53 with mouse double minute 2 homolog (MDM2), a well-known activator of p53 degradation and inhibitor of its DNA transactivation ability (63). Another crucial example of phosphorylation-mediated stability is that of the MYC oncogenic protein. MYC protein stability is regulated by phosphorylation of threonine-58 and serine 62 at the N-terminus (64). Phosphorylation of serine 62 is required for Ras-induced stabilization of MYC, possibly through the action of extracellular-signal regulated kinase (ERK). Phosphorylation at serine 62 also forms a further basis for glycogen synthase kinase 3 (GSK3)-mediated phosphorylation of MYC at threonine-58 that is associated with MYC degradation (64). Since Ras and MYC are critical players for the control of normal cell proliferation, oncogenic events that disrupt this control are associated with altered Ras and MYC signaling. Protein phosphorylation/dephosphorylation also acts as a molecular switch regulating cell survival via altered scaffolding. One example of this is the function of the scaffolding protein, Gravin or A-kinase anchoring protein 12 (AKAP12). AKAP12 is known to interact with protein kinases C and A (PKC/PKA) and localize them to discreet signaling locations. It is known that AKAP12 can sequester cyclin-D1 (CCND1) in the cytoplasm and thereby negatively control cell proliferation (65). However, a specific phosphorylation event at serine 505/serine 517 caused by PKC inhibits the scaffolding activity of AKAP12 for CCND1, leading to nuclear translocation of CCND1 and promotion of G1 to S transition (65). We have recently described that phosphorylation of AKAP12 in hepatic stellate cells alters its ability to scaffold key fibrogenic proteins, heat shock protein 47 (HSP47) and this may be associated with increased collagen expression during hepatic stellate cell activation (66).
Phosphorylation/dephosphorylation-regulated pathways in hepatocellular carcinoma
Mitogen-activated protein kinase/extracellular signal regulated kinases (MAPK/ERK)
Extracellular stimuli such as growth factors, cytokines, mitogens, hormones, and oxidative or heat stress are known to initiate signals on the cell surface by interacting with receptor tyrosine kinases (RTKs), G protein-coupled receptors (GPCRs) or epidermal growth factor receptor (EGFR) (67). These receptors then transmit signals intracellularly by recruiting guanine nucleotide exchange factors to stimulate Ras protein and cause GDP to GTP conversion. This Ras activation event then leads to its binding to Raf proteins that are known MAPK kinase (MAPKKK). This further causes Raf activation via increased kinase activity. MAPKKK activation then causes phosphorylation of MAPKKs and MAPK proteins on threonine and tyrosine residues. MAPK phosphorylation and activation then causes phosphorylation of several downstream targets on serine/threonine residues that causes alterations in a multitude of cellular pathways such as differentiation, gene expression, cell proliferation and apoptosis (68). One of the well-studied MAPKs are ERK1/2. ERK phosphorylation plays an important role in cell proliferation and tumorigenesis. Both its phosphorylation and subsequent kinase activity are dependent on mitogen stimulation and happens in specific phases of the cell cycle. In normal cells, ERK is localized in the cytoplasm where it associates with MAPKKs or with phosphatases (69,70). The MAPK phosphatase 3/dual-specificity phosphatase 6 (MKP3 or DUSP6) is known to localize ERK1/2 in the cytoplasm of unstimulated cells (70). Upon mitogenic stimulation, ERK1/2 get rapidly phosphorylated exhibiting enhanced kinase activity followed by sustenance of moderate kinase activity in the G1 phase of the cell cycle (71). This is followed by nuclear translocation of phospho-ERK that remains functional during late G1 phase and is required for G1 to S transition (72). However, the S phase itself is not dependent upon ERK activation because ERKs are rapidly inactivated after the G1/S transition (72). In the nucleus, ERK is known to phosphorylate several substrates that are responsible for increased cell proliferation. Activated ERK1/2 can phosphorylate the ternary complex factors Elk-1, Sap-1a, and TIF-IA. Phosphorylation of Elk-1 on the C-terminus increases its affinity for the serum response factor and enhances transcription of growth related proteins, such as c-Fos (67). ERK activation can also induce transcription of pro-survival genes (73). ERK can activate the ribosomal S6 family of kinases (RSK1, 2 and 3) and cause them to translocate to the nucleus where they further phosphorylate the TF cAMP response element-binding protein (CREB) at serine 133, which allows recruitment of the co-activators CREB-binding protein (CBP) and p300. This causes CREB-dependent transcription. ERK1 can also directly phosphorylate CBP in vitro and increase the transcriptional activity of CBP (74). ERK-activated RSKs can also induce phosphorylation of the pro-apoptotic protein, BCL2 Associated Agonist of Cell Death (BAD) at a serine 112 residue causes inactivation of its apoptotic function in neuronal cells (75). Given the fact that phospho-ERK is associated with anti-apoptotic and cell survival functions, its role in HCC development and susceptibility has been studied over the past few years. In fact, the multikinase inhibitor, sorafenib that is approved for treatment of advanced HCC, exhibits a clear correlation with the level of phospho-ERK in HCC. HCC cell lines and patient xenografts from HCC specimens that show higher phospho-ERK are more sensitive to the inhibitory effect of sorafenib on growth rate (76). ERK phosphorylation in HCC correlates well with a decrease in expression of MAPK phosphatase 1 (MKP1) or DUSP1 (77). In fact, in human hepatoma cell lines, reactivation of DUSP1 inhibits ERK activation (77). Another important phosphatase that controls the Ras/ERK activation pathway is SHP2 (Src homology 2-containing phosphotyrosine phosphatase 2) that is required to regulate Ras/ERK signaling during tumorigenesis (78). The SUMOylation of SHP2 plays an important role in mediating ERK activation in HCC (78). This will be discussed further in SUMOylation-dependent phosphorylation section of this review.
Phosphatidyl inositol/AK-strain transforming (PI3K/AKT) signaling
The PI3K/AKT signaling pathway is an important survival/proliferative pathway in HCC and is associated with poor prognosis (79). PI3Ks are heterodimeric proteins composed of the p110-catalytic and the p85-regulatory subunit. Cellular stimuli such as mitogens, cytokines cause the p85-regulatory subunit to recruit AKT to the plasma membrane by interacting with phosphorylated motifs on activated receptors. The conversion of phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate (PIP3) by p110-catalytic subunit enhances the kinase activity of phosphatidylinositol 3-kinase-dependent 1, which then phosphorylates and activates AKT at the plasma membrane. In contrast, phosphatase and tensin homolog (PTEN) inactivates PI3K by dephosphorylating PIP3 (80). Hyperactive AKT is one of the mechanisms of acquired resistance to sorafenib in HCC cell lines. An increase in AKT phosphorylation along with upregulation of the p85 subunit is observed in sorafenib-resistant human hepatoma, Huh7 cell line (80). Interestingly, in these resistant cells, downregulation of the phosphatase, PTEN is observed (80).
C-Src signaling
C-SRC is the cellular homolog of the Rous sarcoma virus-transforming (v-Src) gene. It is a non- RTK that plays a role in a variety of signaling pathways associated with cell proliferation. It is known to induce phosphorylation-dependent activation of VEGF, MAPK/ERK and platelet-derived growth factor (PDGF) pathways (81). c-Src activation has been shown to correlate with early stages of HCC phenotypes (82). C-Src is regulated by tyrosine phosphorylation at two sites that result in opposite effects. Tyrosine-416 phosphorylation in the kinase domain of c-Src caused by intermolecular kinase activity of Src itself, induces its enzyme activity whereas phosphorylation of tyrosine-527 by the c-terminal Src kinase (CSK) in the C-terminus inhibits its enzyme activity (83). Increased tyrosine-416 phosphorylation of active c-Src and decreased tyrosine-527 phosphorylation of inactive c-Src is associated with poor patient survival rates in HCC (83). The proline-rich tyrosine kinase 2 (PYK2) that is activated by tyrosine-402 autophosphorylation, has been shown to interact with c-Src and promote its activation through tyrosine-416 phosphorylation in HCC cell lines (84). Interestingly, this autophosphorylation activity and SRC-recruitment activity of PYK2 has recently been shown to be dependent on SUMOylation (85). This signaling is further discussed under the SUMOylation-dependent phosphorylation section of this review.
SUMOylation-dependent phosphorylation/phosphorylation-dependent SUMOylation
The overall goal of this work is to review the cross-talk between SUMOylation and phosphorylation pathways during HCC development. Although there is substantial literature that suggests dependence of phosphorylation on SUMOylation in HCC targets, there is no literature on phosphorylation-dependent SUMOylated targets in HCC. Phosphorylation-dependent SUMOylation modifications (PDSMs) are described in general (86). For example, phosphorylation has been shown to reduce the SUMOylation of the ELK1, c-Fos and c-Jun TFs and increase the transcriptional activity of the nuclear factor erythroid-derived 2 (NF-E2) protein (87). Examples of SUMOylation-dependent phosphorylation in HCC are the Ras/ERK pathway and the C-Src signaling pathway. The activation of the Ras/ERK pathway is controlled by the activity of SHP2 protein during tumorigenesis (78). A major SUMO-acceptor site, lysine-590 in SHP2 was recently identified to be SUMOylated by SUMO-1. This SUMOylation modification enhanced epidermal growth factor (EGF)-mediated ERK phosphorylation and activation in HCC cell lines. The mechanism by which SUMO-SHP2 facilitated ERK activation and HCC tumor growth was by enhancement of binding of SHP2 to the Grb2-associated binder-1 (GAB1) protein (78). Another interesting interaction between SUMOylation and phosphorylation exists for the c-Src protein and PYK2. PYK2 gets autophosphorylated and in this form it interacts with and induces the tyrosine-416 activity of c-Src in HCC cells (84). Recently, global SUMO-proteome analysis revealed that several cytosolic non- RTKs including PYK2 were SUMOylation targets (85). Further analysis of SUMO-PYK2 revealed that this adduct caused rapid Src phosphorylation at tyrosine-416 compared to unSUMOylated PYK2 or a SUMO-mutant of PYK2 in vitro (85). Another important correlation exists between PYK2 and ERK1/2 phosphorylation and activation in HCC (84). This enhancement of ERK activity was also induced significantly by PYK2 SUMOylation and inhibited by a PYK2 SUMO mutant (85). The overall data imply a correlation between enhanced PYK2 autophosphorylation, SUMOylation and C-Src signaling as well as enhanced ERK activation in HCC. The E2 enzyme UBC9 is also phosphorylated in human HCC and during inflammatory signaling in Kupffer cells of the liver (12,54). Serine 71 phosphorylation of UBC9 appears to favor its stability in cancer cell lines (12). Phosphorylation of UBC9 also promotes inflammatory signaling in Kupffer cells of the liver (54). Since SUMOylation is also higher in HCC, it appears that phosphorylation of UBC9 may drive enhanced SUMOylation in this disease (12).
Acknowledgements
Funding: This work was supported by NIH grant K01AA022372 (ML Tomasi)
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Alison MR. Liver stem cells: implication for hepatocarcinogenesis. Stem Cell Rev 2005;1:253-60. [Crossref] [PubMed]
- Bugianesi E. Non-alcoholic steatohepatitis and cancer. Clin Liver Dis 2007;11:191-207. [Crossref] [PubMed]
- Krueger KE, Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics 2006;5:1799-810. [Crossref] [PubMed]
- Duan G, Walther D. The Roles of Post-translational Modifications in the Context of Protein Interaction Networks. PLoS Comput Biol 2015;11:e1004049. [Crossref] [PubMed]
- Prasad S, Pal PK. SUMOylation: One small modification for proteins, multiple giant problems for mankind. Mov Disord 2018;33:403. [Crossref] [PubMed]
- Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 2013;82:357-85. [Crossref] [PubMed]
- Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 2010;428:133-45. [Crossref] [PubMed]
- Hay RT. SUMO-specific proteases: a twist in the tail. Trends Cell Biol 2007;17:370-6. [Crossref] [PubMed]
- Liu B, Shuai K. Summon SUMO to wrestle with inflammation. Mol Cell 2009;35:731-2. [Crossref] [PubMed]
- Guo WH, Yuan LH, Xiao ZH, et al. Overexpression of SUMO-1 in hepatocellular carcinoma: a latent target for diagnosis and therapy of hepatoma. J Cancer Res Clin Oncol 2011;137:533-41. [Crossref] [PubMed]
- Lee JS, Thorgeirsson SS. Genome-scale profiling of gene expression in hepatocellular carcinoma: classification, survival prediction, and identification of therapeutic targets. Gastroenterology 2004;127:S51-5. [Crossref] [PubMed]
- Tomasi ML, Tomasi I, Ramani K, et al. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology 2012;56:982-93. [Crossref] [PubMed]
- Jiang QF, Tian YW, Shen Q, et al. SENP2 regulated the stability of beta-catenin through WWOX in hepatocellular carcinoma cell. Tumour Biol 2014;35:9677-82. [Crossref] [PubMed]
- Shen HJ, Zhu HY, Yang C, et al. SENP2 regulates hepatocellular carcinoma cell growth by modulating the stability of beta-catenin. Asian Pac J Cancer Prev 2012;13:3583-7. [Crossref] [PubMed]
- Qin Y, Bao H, Pan Y, et al. SUMOylation alterations are associated with multidrug resistance in hepatocellular carcinoma. Mol Med Rep 2014;9:877-81. [Crossref] [PubMed]
- Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev 2005;15:536-41. [Crossref] [PubMed]
- Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans 2007;35:1389-92. [Crossref] [PubMed]
- Rosonina E, Akhter A, Dou Y, et al. Regulation of transcription factors by sumoylation. Transcription 2017;8:220-31. [Crossref] [PubMed]
- Bies J, Markus J, Wolff L. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J Biol Chem 2002;277:8999-9009. [Crossref] [PubMed]
- Muller S, Berger M, Lehembre F, et al. c-Jun and p53 activity is modulated by SUMO-1 modification. J Biol Chem 2000;275:13321-9. [Crossref] [PubMed]
- Mato JM, Alvarez L, Ortiz P, et al. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther 1997;73:265-80. [Crossref] [PubMed]
- Lu Z, Wu H, Mo YY. Regulation of bcl-2 expression by Ubc9. Exp Cell Res 2006;312:1865-75. [Crossref] [PubMed]
- Yang H, Sadda MR, Yu V, et al. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor alpha. Role of NF-kappa B and AP-1. J Biol Chem 2003;278:50887-96. [Crossref] [PubMed]
- Hayes JD, Chanas SA, Henderson CJ, et al. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochem Soc Trans 2000;28:33-41. [Crossref] [PubMed]
- Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med 2009;47:1304-9. [Crossref] [PubMed]
- Ramani K, Tomasi ML, Yang H, et al. Mechanism and significance of changes in glutamate-cysteine ligase expression during hepatic fibrogenesis. J Biol Chem 2012;287:36341-55. [Crossref] [PubMed]
- Tomasi ML, Ryoo M, Yang H, et al. Molecular mechanisms of lipopolysaccharide-mediated inhibition of glutathione synthesis in mice. Free Radic Biol Med 2014;68:148-58. [Crossref] [PubMed]
- Comel A, Sorrentino G, Capaci V, et al. The cytoplasmic side of p53's oncosuppressive activities. FEBS Lett 2014;588:2600-9. [Crossref] [PubMed]
- Hock A, Vousden KH. Regulation of the p53 pathway by ubiquitin and related proteins. Int J Biochem Cell Biol 2010;42:1618-21. [Crossref] [PubMed]
- Lu X, Yi J. SUMO-1 enhancing the p53-induced HepG2 cell apoptosis. J Huazhong Univ Sci Technolog Med Sci 2005;25:289-91. [Crossref] [PubMed]
- Medrano EE. Repression of TGF-β signaling by the oncogenic protein SKI in human melanomas. Consequences for proliferation, survival, and metastasis. Oncogene 2003;22:3123-9. [Crossref] [PubMed]
- Xu W, Angelis K, Danielpour D, et al. Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor. Proc Natl Acad Sci U S A 2000;97:5924-9. [Crossref] [PubMed]
- Tokitou F, Nomura T, Khan MM, et al. Viral ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J Biol Chem 1999;274:4485-8. [Crossref] [PubMed]
- Ding B, Sun Y, Huang J. Overexpression of SKI oncoprotein leads to p53 degradation through regulation of MDM2 protein sumoylation. J Biol Chem 2012;287:14621-30. [Crossref] [PubMed]
- Knipscheer P, Flotho A, Klug H, et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 2008;31:371-82. [Crossref] [PubMed]
- Semenza GL. HIF-1 and human disease: one highly involved factor. Genes Dev 2000;14:1983-91. [PubMed]
- Li J, Xu Y, Long XD, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell 2014;25:118-31. [Crossref] [PubMed]
- Jiao HK, Xu Y, Li J, et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis 2015;6:e1689. [Crossref] [PubMed]
- Klenk C, Humrich J, Quitterer U, et al. SUMO-1 controls the protein stability and the biological function of phosducin. J Biol Chem 2006;281:8357-64. [Crossref] [PubMed]
- Müller S, Hoege C, Pyrowolakis G, et al. Nat Rev Mol Cell Biol 2001;2:202-10. [Crossref] [PubMed]
- Nait Achour T, Sentis S, Teyssier C, et al. Transcriptional repression of estrogen receptor alpha signaling by Senp2 in breast cancer cells. Mol Endocrinol 2014;28:183-96. [Crossref] [PubMed]
- Del Mare S, Kurek KC, Stein GS, et al. Role of the WWOX tumor suppressor gene in bone homeostasis and the pathogenesis of osteosarcoma. Am J Cancer Res 2011;1:585-94. [PubMed]
- Bouteille N, Driouch K, Hage PE, et al. Inhibition of the Wnt/beta-catenin pathway by the WWOX tumor suppressor protein. Oncogene 2009;28:2569-80. [Crossref] [PubMed]
- Zhang L, Rubins NE, Ahima RS, et al. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab 2005;2:141-8. [Crossref] [PubMed]
- Qian X, Costa RH. Analysis of hepatocyte nuclear factor-3 beta protein domains required for transcriptional activation and nuclear targeting. Nucleic Acids Res 1995;23:1184-91. [Crossref] [PubMed]
- Liu M, Lee DF, Chen CT, et al. IKKα activation of NOTCH links tumorigenesis via FOXA2 suppression. Mol Cell 2012;45:171-84. [Crossref] [PubMed]
- Belaguli NS, Zhang M, Brunicardi FC, et al. Forkhead Box Protein A2 (FOXA2) Protein Stability and Activity Are Regulated by Sumoylation. PLoS One 2012;7:e48019. [Crossref] [PubMed]
- Chuma M, Sakamoto N, Nakai A, et al. Heat shock factor 1 accelerates hepatocellular carcinoma development by activating nuclear factor-κB/mitogen-activated protein kinase. Carcinogenesis 2014;35:272-81. [Crossref] [PubMed]
- Liu Y, Lou G, Wu W, et al. Interferon-α sensitizes HBx-expressing hepatocarcinoma cells to chemotherapeutic drugs through inhibition of HBx-mediated NF-κB activation. Virol J 2013;10:168. [Crossref] [PubMed]
- Wang H, Wang X, Li X, et al. CD68(+)HLA-DR(+) M1-like macrophages promote motility of HCC cells via NF-κB/FAK pathway. Cancer Lett 2014;345:91-9. [Crossref] [PubMed]
- Aravindan N, Aravindan S, Pandian V, et al. Acquired tumor cell radiation resistance at the treatment site is mediated through radiation-orchestrated intercellular communication. Int J Radiat Oncol Biol Phys 2014;88:677-85. [Crossref] [PubMed]
- Ordoñez R, Carbajo-Pescador S, Prieto-Dominguez N, et al. Inhibition of matrix metalloproteinase-9 and nuclear factor kappa B contribute to melatonin prevention of motility and invasiveness in HepG2 liver cancer cells. J Pineal Res 2014;56:20-30. [Crossref] [PubMed]
- Gilmore TD, Wolenski FS. NF-κB: where did it come from and why? Immunol Rev 2012;246:14-35. [Crossref] [PubMed]
- Tomasi ML, Ramani K, Ryoo M. Ubiquitin-Conjugating Enzyme 9 Phosphorylation as a Novel Mechanism for Potentiation of the Inflammatory Response. Am J Pathol 2016;186:2326-36. [Crossref] [PubMed]
- Liu Y, Bridges R, Wortham A, et al. NF-κB repression by PIAS3 mediated RelA SUMOylation. PLoS One 2012;7:e37636. [Crossref] [PubMed]
- Frank S, Peters MA, Wehmeyer C, et al. Regulation of matrixmetalloproteinase-3 and matrixmetalloproteinase-13 by SUMO-2/3 through the transcription factor NF-κB. Ann Rheum Dis 2013;72:1874-81. [Crossref] [PubMed]
- Liu J, Sha M, Wang Q, et al. Small ubiquitin-related modifier 2/3 interacts with p65 and stabilizes it in the cytoplasm in HBV-associated hepatocellular carcinoma. BMC Cancer 2015;15:675. [Crossref] [PubMed]
- Kim EM, Lee HH, Kim SH, et al. The mouse small ubiquitin-like modifier-2 (SUMO-2) inhibits interleukin-12 (IL-12) production in mature dendritic cells by blocking the translocation of the p65 subunit of NFκB into the nucleus. Mol Immunol 2011;48:2189-97. [Crossref] [PubMed]
- Liu J, Tao X, Zhang J, et al. Small ubiquitin-related modifier 1 is involved in hepatocellular carcinoma progression via mediating p65 nuclear translocation. Oncotarget 2016;7:22206-18. [PubMed]
- Ardito F, Giuliani M, Perrone D, et al. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy. Int J Mol Med 2017;40:271-80. [Crossref] [PubMed]
- Roskoski R Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res 2012;66:105-43. [Crossref] [PubMed]
- Ashcroft M, Kubbutat MH, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol 1999;19:1751-8. [Crossref] [PubMed]
- Ashcroft M, Vousden KH. Regulation of p53 stability. Oncogene 1999;18:7637-43. B. [Crossref] [PubMed]
- Sears R, Nuckolls F, Haura E, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000;14:2501-14. [Crossref] [PubMed]
- Lin X, Nelson P, Gelman IH. SSeCKS, a major protein kinase C substrate with tumor suppressor activity, regulates G(1)–>S progression by controlling the expression and cellular compartmentalization of cyclin D. Mol Cell Biol 2000;20:7259-72. [Crossref] [PubMed]
- Ramani K, Tomasi ML, Berlind J, et al. Role of A-Kinase Anchoring Protein Phosphorylation in Alcohol-Induced Liver Injury and Hepatic Stellate Cell Activation. Am J Pathol 2018;188:640-55. [Crossref] [PubMed]
- Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle 2009;8:1168-75. [Crossref] [PubMed]
- Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci 1993;18:128-31. [Crossref] [PubMed]
- Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. Embo J 1997;16:1901-8. [Crossref] [PubMed]
- Karlsson M, Mathers J, Dickinson RJ, et al. Both nuclear-cytoplasmic shuttling of the dual specificity phosphatase MKP-3 and its ability to anchor MAP kinase in the cytoplasm are mediated by a conserved nuclear export signal. J Biol Chem 2004;279:41882-91. [Crossref] [PubMed]
- Meloche S. Cell cycle reentry of mammalian fibroblasts is accompanied by the sustained activation of p44mapk and p42mapk isoforms in the G1 phase and their inactivation at the G1/S transition. J Cell Physiol 1995;163:577-88. [Crossref] [PubMed]
- Yamamoto T, Ebisuya M, Ashida F, et al. Continuous ERK activation downregulates antiproliferative genes throughout G1 phase to allow cell-cycle progression. Curr Biol 2006;16:1171-82. [Crossref] [PubMed]
- Lu Z, Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life 2006;58:621-31. [Crossref] [PubMed]
- Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem Biophys Res Commun 1996;228:831-37. [Crossref] [PubMed]
- Hayakawa J, Ohmichi M, Kurachi H, et al. Inhibition of BAD phosphorylation either at serine 112 via extracellular signal-regulated protein kinase cascade or at serine 136 via Akt cascade sensitizes human ovarian cancer cells to cisplatin. Cancer Res 2000;60:5988-94. [PubMed]
- Liang Y, Chen J, Yu Q, et al. Phosphorylated ERK is a potential prognostic biomarker for Sorafenib response in hepatocellular carcinoma. Cancer Med 2017;6:2787-95. [Crossref] [PubMed]
- Calvisi DF, Pinna F, Meloni F, et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res 2008;68:4192-200. [Crossref] [PubMed]
- Deng R, Zhao X, Qu Y, et al. Shp2 SUMOylation promotes ERK activation and hepatocellular carcinoma development. Oncotarget 2015;6:9355-69. [Crossref] [PubMed]
- Nakanishi K, Sakamoto M, Yamasaki S, et al. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer 2005;103:307-12. [Crossref] [PubMed]
- Chen KF, Chen HL, Tai WT, et al. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther 2011;337:155-61. [Crossref] [PubMed]
- Liu W, Yue F, Zheng M, et al. The proto-oncogene c-Src and its downstream signaling pathways are inhibited by the metastasis suppressor, NDRG1. Oncotarget 2015;6:8851-74. [PubMed]
- Ito Y, Kawakatsu H, Takeda T, et al. Activation of c-Src gene product in hepatocellular carcinoma is highly correlated with the indices of early stage phenotype. J Hepatol 2001;35:68-73. [Crossref] [PubMed]
- Chen ML, Chai CY, Yeh KT, et al. Crosstalk between activated and inactivated c-Src in hepatocellular carcinoma. Dis Markers 2011;30:325-33. [Crossref] [PubMed]
- Sun CK, Man K, Ng KT, et al. Proline-rich tyrosine kinase 2 (Pyk2) promotes proliferation and invasiveness of hepatocellular carcinoma cells through c-Src/ERK activation. Carcinogenesis 2008;29:2096-105. [Crossref] [PubMed]
- Uzoma I, Hu J, Cox E, et al. Global Identification of SUMO Substrates Reveals Crosstalk between SUMOylation and Phosphorylation Promotes Cell Migration. Mol Cell Proteomics 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Hietakangas V, Anckar J, Blomster HA, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 2006;103:45-50. [Crossref] [PubMed]
- Su YF, Shyu YC, Shen CK, et al. Phosphorylation-dependent SUMOylation of the transcription factor NF-E2. PLoS One 2012;7:e44608. [Crossref] [PubMed]
Cite this article as: Tomasi ML, Ramani K. SUMOylation and phosphorylation cross-talk in hepatocellular carcinoma. Transl Gastroenterol Hepatol 2018;3:20.


